Family: Betaflexiviridae
Chapter Version: ICTV Ninth Report; 2009 Taxonomy Release
Distinguishing features
The family contains viruses infecting plants which share a distinct lineage of alphavirus-like replication proteins.
Virion properties
Morphology
Virions are flexuous filaments, usually 12–13 nm in diameter (range 10–15 nm) and from 600 to over 1000 nm in length, depending on the genus. They have helical symmetry with a pitch of about 3.4 nm (range 3.3–3.7 nm) and in some genera there is clearly visible cross-banding.
Physicochemical and physical properties
Virions sediment as single (or occasionally two very close) bands with an S20,w of 92–176S, depending on the genus.
Nucleic acid
Virions contain a single molecule of linear ssRNA of about 5.9–9.0 kb which is 5–6% by weight of the virion. The RNA is capped (or probably capped) at the 5’ terminus with m7G and has a polyadenylated tract at the 3’ terminus. In the genus Carlavirus some viruses have two subgenomic RNAs (sgRNAs) of 2.1–3.3 kb and 1.3–1.6 kb, which are possibly encapsidated in shorter particles.
Proteins
The viral capsid of all members is composed of a single polypeptide ranging in size from 18 to 44 kDa.
Lipids
None reported.
Carbohydrates
None reported.
Genome organization and replication
The number of genes is between three and six depending upon the genus (Figure 1) but, in all species, the ORF1-encoded product, which follows a short 5’-UTR sequence, has homologies with polymerase proteins of the “alphavirus-like” supergroup of RNA viruses. This protein (190–250 kDa) contains the conserved domains for methyltransferase (Mtr), helicase (Hel) and RNA-dependent RNA polymerase (RdRp) activity. Most members also have AlkB and papain-like protease (P-Pro) domains between the Mtr and Hel. Smaller ORFs encode the proteins involved in cell-to-cell movement, either a single MP of the “30K” superfamily (Capillovirus, Citrivirus, Trichovirus, Vitivirus) or a “triple gene block” (TGB) (remaining genera and viruses). These are usually located following (3’-proximal to) the polymerase but in capillovirus genomes the MP ORF2 is nested within the ORF1 and in vitiviruses an extra ORF is present between the polymerase and MP genes. The CP gene always follows the MP(s) and in some genera (Carlavirus, Vitivirus and some trichoviruses) a final ORF encodes a protein with a zinc binding finger motif and the ability to bind nucleic acids. In vitiviruses, this small protein has been shown to have RNA silencing suppressor activity. ORFs downstream of the polymerase are translated from 3’-terminal sgRNAs that can often be found in infected tissue. In some viruses, notably in the genera Citrivirus, Vitivirus and Trichovirus, nested sets of 5’-terminal sgRNAs and their associated dsRNAs can also be detected. Replication is (or is presumed to be) cytoplasmic and the product of ORF1 is the only virus-encoded protein known to be involved.
Antigenic properties
Virions are highly immunogenic in members of the genus Carlavirus but those of other genera are only moderate to poor antigens. Within (but not usually between) genera, some viruses are serologically related.
Biological properties
Members have been reported from a diverse range of plant species but the host range of individual members is usually limited. With the exception of most members of the genus Carlavirus, natural infections are mostly or exclusively of woody hosts. Many of the viruses have relatively mild effects on their host. All species can be transmitted by mechanical inoculation, although some with difficulty. Many of the viruses have no known invertebrate or fungus vectors; however some trichoviruses are known to be mite-borne, most carlaviruses are transmitted naturally by aphids in the non-persistent manner and a range of vectors (pseudococcid mealybugs, scale insects and aphids) have been reported for different vitiviruses. Aggregates of virus particles accumulate in the cytoplasm. Many carlaviruses induce the formation of ovoid or irregularly shaped inclusions but otherwise there are usually no specific cytopathic structures.
Species and genus demarcation criteria in the family
Genera are distinguished by various features of genome organization and the natural mode of transmission. These are summarized in Table 1. Throughout the family, isolates of different species should have less than about 72% nt identity (or 80% aa identity) between their respective CP or polymerase genes. Viruses from different genera usually have less than about 45% nt identity in these genes.
Table 1 Distinguishing properties of genera in the family Betaflexiviridae
| Genus | Virion length (nm) | ORFs | Repa | MP(s)b | CPc |
| Capillovirus | 640–700 | 2 | 210–245 | 30K | 25–27 |
| Carlavirus | 610–700 | 6 | 215–225 | TGB | 32–36 |
| Citrivirus | 960 | 3 | 227 | 30K | 41 |
| Foveavirus | 800+ | 5 | 230–250 | TGB | 28–44 |
| Trichovirus | 640–890 | 3 or 4 | 215–220 | 30K | 21–24 |
| Vitivirus | 725–785 | 5 | 190–200 | 30K | 18–22 |
a Rep, Replication protein size (kDa).
b MP, Movement protein either of the “30K” superfamily or a triple gene block (TGB).
c CP, Coat protein size (kDa).
Genus Capillovirus
Type species Apple stem grooving virus
Distinguishing features
Capilloviruses have a distinctive genomic organization, with two ORFs encoding a large replication-associated protein fused with the coat protein and (as a nested ORF) a putative movement protein. The MP and CP are expressed from subgenomic RNAs. No vectors are known. Virions have prominent cross-banding.
Virion properties
Morphology
Virions are flexuous filaments, 640–700×12 nm, constructed from helically arranged protein subunits in a primary helix with a pitch of 3.4 nm and between 9 and 10 subunits per turn with prominent cross-banding (see Figure 1 above).
Physicochemical and physical properties
The S20,w of particles is about 112S, isoelectric point is about pH 4.3 at ionic strength 0.1 M, and electrophoretic mobility is 10.3 and 6.5×10−5 cm−2 sec−1 volt−1, at pH 7.0 and 6.0 respectively (ionic strength 0.1 M; data for apple stem grooving virus).
Nucleic acid
Virions contain linear positive sense ssRNA, 6.5–7.4 kb in size, constituting about 5%, by weight, of virions. The RNA is polyadenylated at its 3’ end. Isolates of the species Apple stem grooving virus from different hosts show wide variations in the sequence of a 284 aa region of ORF1-encoded protein, between the polymerase and CP domains.
Proteins
Virions are composed of a single 24–27 kDa protein.
Genome organization and replication
The genomic RNA of all sequenced viruses has the same organization, and two ORFs (Figure 2). ORF1 encodes a 240–266 kDa protein followed by a UTR of 140–300 nt upstream of the 3’-poly(A) tail. ORF2 is nested within ORF1 near its 3’ end, and encodes a 36–52 kDa protein. Although the CP cistron is located in the C-terminal end of ORF1, and ORF2 is nested within ORF1, the strategy of expression of both CP and putative MP may be based on sgRNA production, as suggested by the analysis of dsRNA patterns from infected tissues. dsRNAs of the type member consist of five major bands with sizes of approximately 6.5, 5.5, 4.5, 2.0 and 1.0 kbp. The 6.5 kbp species probably represents the double stranded form of the full-length genome, and the 2.0 and the 1.0 kbp species may be the double-stranded forms of sgRNAs that code for the putative MP and the CP, respectively. Replication is likely to occur in the cytoplasm, in which virus particles accumulate in discrete bundles.
Antigenic properties
Virions are moderately antigenic. There are no serological relationships between species.
Biological properties
Host range
Apple stem grooving virus (ASGV) is pathogenic to pome fruits and citrus and induces stock/scion incompatibility, i.e. top-working disease of apple and bud union crease syndrome of citrus. It also infects lily. Cherry virus A (CVA) is frequently found in sweet and sour cherry (and less frequently in other Prunus hosts) but no disease has been associated with it.
Transmission
No vectors are known. ASGV was transmitted through seed to progeny seedlings of Chenopodium quinoa, and lily. ASGV, CVA, and Nandina stem pitting virus (NSPV) are transmitted by grafting. NSPV has not been transmitted by sap inoculation, but by slashing stems with a partially purified virus preparation.
Geographical distribution
Geographical distribution ranges from wide to restricted according to the virus. ASGV has been recorded from most areas where apples are grown, and is widespread in citrus in China, Japan, United States, Australia and South Africa. CVA is widespread and probably occurs worldwide in cherry hosts. NSPV is found only in the United States.
Cytopathic effects
No distinct cytological alterations have been observed in infected cells. Virus particles occur in bundles in mesophyll and phloem parenchyma cells, but not in the epidermis and sieve elements.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Natural host range.
- Serological specificity (all known species are serologically unrelated).
- Less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
List of species in the genus Capillovirus
| Apple stem grooving virus |
|
|
| Apple stem grooving virus-P-209 | [D14995=NC_001749] | (ASGV-P209) |
| Citrus tatter leaf virus-lily | [D16681] | (CTLV-L) |
| Cherry virus A |
|
|
| Cherry virus A-Germany | [X82547=NC_003689] | (CVA-DE) |
Species names are in italic script; names of isolates and strains are in roman script. Sequence accessions [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Capillovirus but have not been approved as species
| Nandina stem pitting virus |
| (NSPV) |
Genus Carlavirus
Type species Carnation latent virus
Distinguishing features
Carlaviruses have six ORFs, including a TGB, and are insect-transmitted.
Virion properties
Morphology
Virions are slightly flexuous filaments, 610–700 nm in length and 12–15 nm in diameter (Figure 3). They have helical symmetry with a pitch of about 3.4 nm.
Physicochemical and physical properties
Virion Mr is about 60×106, with a nucleic acid content of about 6%. Virion S20,w is 147–176S, and the buoyant density in CsCl solutions is 1.3 g cm−3.
Nucleic acid
Virions contain a single molecule of linear ssRNA that has a size range of 7.4–7.7 kb when estimated by agarose gel analysis, although full-length sequence analysis suggests that genome sizes are in the 8.3–8.7 kb range. Some species also have two sgRNAs of 2.1–3.3 kb and 1.3–1.6 kb, which are possibly encapsidated in shorter particles. The genomic RNAs have a 3’-poly(A) tract and a 5’-cap.
Genome organization and replication
There are typically six ORFs with short UTRs at the 5’ and 3’ termini. In potato virus M (Figure 4), ORF1 encodes a polypeptide of 223 kDa that is the viral replicase; ORFs 2, 3 and 4 form the triple gene block and encode polypeptides of 25, 12 and 7 kDa which facilitate virus movement. ORF5 encodes the 34 kDa CP and overlaps ORF6, which encodes a cysteine-rich protein of 11–16 kDa. The function of the 11–16 kDa polypeptide has yet to be determined, but its ability to bind nucleic acid indicates that it may facilitate aphid transmission or be involved in host gene transcription/gene silencing and/or viral RNA replication.
Only ORF1 is translated from the full length genomic RNA. With blueberry scorch virus and probably other carlaviruses the product of ORF1 is proteolytically processed by a papain-like proteinase activity, with about 30–40 kDa being removed. The 3’-terminal ORFs appear to be translated from two sgRNAs that can be found in infected tissue, and, for some viruses, can be detected in purified virus preparations. The 5’-untranslated leader sequence of the genomic RNA and the sgRNA for the CP of potato virus S (PVS) have both been shown to act as efficient enhancers of translation.
Antigenic properties
Carlavirus virions are good immunogens. Some species are serologically interrelated, but others are apparently distinct.
Biological properties
Host range
Individual viruses have restricted natural host ranges, but some can infect a wide range of experimental hosts.
Transmission
Most species are transmitted naturally by aphids in the non-persistent manner; cowpea mild mottle virus (CPMMV) is transmitted by whiteflies (Bemisia tabaci), pea streak virus, red clover vein mosaic virus and CPMMV are seedborne in their leguminous hosts. All are mechanically transmissible; some (e.g. carnation latent virus and PVS) are sufficiently infectious to be so transmitted this way in the field.
Geographical distribution
The geographical distribution of many species is restricted, but those infecting vegetatively-propagated crops are usually widely distributed, presumably due to inadvertent dissemination in vegetative propagules. Most species commonly occur in temperate climates, but CPMMV is restricted to tropical and sub-tropical regions.
Cytopathic effects
Virions of aphid-borne species are scattered throughout the cytoplasm or occur in membrane-associated bundle-like or plate-like aggregates. Many species also induce the formation of ovoid or irregularly shaped inclusions that appear in the light microscope as vacuolate bodies; these consist of aggregates of virus particles, mitochondria, endoplasmic reticulum and lipid globules. The particles of CPMMV, the whitefly-transmitted carlavirus, also occur in aggregates in cytoplasm; those of most, but not all, strains of CPMMV form brush-like inclusions.
Species demarcation criteria in the genus
Each distinct species usually has a specific natural host range. Distinct species do not cross-protect in infected common host plant species. Distinct species are readily differentiated by serological procedures; strains of individual species are often distinguishable in reactions with polyclonal antisera, but more readily so with monoclonal antibodies. Distinct species have less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
List of species in the genus Carlavirus
| Aconitum latent virus |
|
|
| Aconitum latent virus-Japan:D | [AB051848=NC_002795] | (AcLV-D) |
| American hop latent virus |
|
|
| American hop latent virus-USA:Washington State |
| (AHLV-WA) |
| Blueberry scorch virus |
|
|
| Blueberry scorch virus-NJ-2 | [L25658=NC_003499] | (BlScV-nj2) |
| Cactus virus 2 |
|
|
| Cactus virus 2-Germany |
| (CV-2-DE) |
| Caper latent virus |
|
|
| Caper latent virus-Italy |
| (CapLV-IT) |
| Carnation latent virus |
|
|
| Carnation latent virus-United Kingdom | [AJ010697*] | (CLV-UK) |
| Chrysanthemum virus B |
|
|
| Chrysanthemum virus B-Japan:Showa | [AB245142] | (CVB-S) |
| Cole latent virus |
|
|
| Cole latent virus-Brazil | [AY340584*] | (CoLV-BR) |
| Coleus vein necrosis virus |
|
|
| Coleus vein necrosis virus-USA | [EF527260=NC_009764] | (CVNV-USA) |
| Cowpea mild mottle virus |
|
|
| Cowpea mild mottle virus-M | [AF024629*] | (CPMMV-M) |
| Dandelion latent virus |
|
|
| Dandelion latent virus-Canada:British Colombia |
| (DaLV-BC) |
| Daphne virus S |
|
|
| Daphne virus S-type strain: K | [AJ620300=NC_008020] | (DVS-K) |
| Elderberry symptomless virus |
|
|
| Elderberry symptomless virus-United Kingdom |
| (ElSLV-UK) |
| Garlic common latent virus |
|
|
| Garlic common latent virus-Germany | [AB004805*] | (GarCLV-DE) |
| Helenium virus S |
|
|
| Helenium virus S-Germany | [D10454*] | (HVS-DE) |
| Helleborus net necrosis virus |
|
|
| Helleborus net necrosis virus-G5 | [FJ196835=NC_012038] | (HNNV-G5) |
| Honeysuckle latent virus |
|
|
| Honeysuckle latent virus-United Kingdom |
| (HnLV-UK) |
| Hop latent virus |
|
|
| Hop latent virus-Japan | [AB032469=NC_002552] | (HpLV-JA) |
| Hop mosaic virus |
|
|
| Hop mosaic virus-Australia | [EU527979=NC_010538] | (HpMV-AUS) |
| Hydrangea latent virus |
|
|
| Hydrangea latent virus-USA |
| (HdLV-USA) |
| Kalanchoë latent virus |
|
|
| Kalanchoë latent virus-PV-0290B | [FJ531634=NC_013006] | (KLV-PV0290B) |
| Ligustrum necrotic ringspot virus |
|
|
| Ligustrum necrotic ringspot virus-USA | [EU074853=NC_010305] | (LNRSV-USA) |
| Lilac mottle virus |
|
|
| Lilac mottle virus-USA |
| (LiMoV-USA) |
| Lily symptomless virus |
|
|
| Lily symptomless virus-South Korea | [AJ516059=NC_005138] | (LSV-Kor) |
| Melon yellowing-associated virus |
|
|
| Melon yellowing-associated virus-Bessa | [AB510477*] | (MYaV-Bessa) |
| Mulberry latent virus |
|
|
| Mulberry latent virus-Japan |
| (MLV-JP) |
| Muskmelon vein necrosis virus |
|
|
| Muskmelon vein necrosis virus-USA:California |
| (MuVNV-CAL) |
| Narcissus common latent virus |
|
|
| Narcissus common latent virus-Zhangzhou | [AM158439=NC_008266] | (NCLV-ZZ) |
| Narcissus symptomless virus |
|
|
| Narcissus symptomless virus-Hangzhou | [AM182569=NC_008552] | (NSV-HZ) |
| Nerine latent virus |
|
|
| (Hippeastrum latent virus) |
|
|
| Nerine latent virus-Taiwan | [DQ098905=NC_011540] | (NeLV-TW) |
| Passiflora latent virus |
|
|
| Passiflora latent virus-Israel | [DQ455582=NC_008292] | (PLV-IS) |
| Pea streak virus |
|
|
| Pea streak virus-ATCCPV-87 | [AF354652*] | (PeSV-ATCCPV87) |
| Poplar mosaic virus |
|
|
| Poplar mosaic virus-PV-0341 | [AY505475=NC_005343] | (PopMV-PV0341) |
| Potato latent virus |
|
|
| Potato latent virus-Canada | [EU433397=NC_011525] | (PotLV-CAN) |
| Potato virus M |
|
|
| Potato virus M-Russian wild type | [D14449=NC_001361] | (PVM-RU) |
| Potato virus P |
|
|
| Potato virus P-Brazil | [EU338239] | (PVP-BRZ) |
| Potato rough dwarf virus | [EU020009=NC_009759] | (PRDV) |
| Potato virus S |
|
|
| Potato virus S-Leona | [AJ863509=NC_007289] | (PVS-Leona) |
| Red clover vein mosaic virus |
|
|
| Red clover vein mosaic virus-Washington | [FJ685618=NC_012210] | (RCVMV-Washington) |
| Shallot latent virus |
|
|
| Shallot latent virus-YH1 | [AJ292226=NC_003557] | (SLV-YH1) |
| Sint-Jan’s onion latent virus |
|
|
| Sint-Jan’s onion latent virus-Netherlands |
| (SJOLV-NL) |
| Strawberry pseudo mild yellow edge virus |
|
|
| Strawberry pseudo mild yellow edge virus-USA |
| (SPMYEV-USA) |
| Sweet potato chlorotic fleck virus |
|
|
| Sweet potato chlorotic fleck virus-Uganda | [AY461421=NC_006550] | (SPCFV-UG) |
| Verbena latent virus |
|
|
| Verbena latent virus-Israel | [AF271218*] | (VeLV-IS) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Carlavirus but have not been approved as species
| Arracacha latent virus |
| (ALV) |
| Artichoke latent virus M |
| (ArLVM) |
| Artichoke latent virus S |
| (ArLVS) |
| Butterbur mosaic virus | [AB517596= NC_013527] | (ButMV) |
| Cardamine latent virus |
| (CaLV) |
| Carrot virus S | [EU881919*] | (CarVS) |
| Helleborus mosaic virus | [FJ196838*] | (HeMV) |
| Hydrangea chlorotic mottle virus | [EU754720= NC_012869] | (HCMoV) |
| Phlox virus B | [EU162589= NC_009991] | (PhlVB) |
| Phlox virus M | [EF507476*] | (PhlVM) |
| Phlox virus S | [EF492068= NC_009383] | (PhlVS) |
| Sedum latent virus | [FJ560901*] | (SeLV) |
* Sequences do not comprise the complete genome.
Genus Citrivirus
Type species Citrus leaf blotch virus
Distinguishing features
This genus consists of a single species. There are three ORFs, similar to trichoviruses, but it is distinct from them in phylogenetic analyses, has longer virions and a much larger coat protein that more closely resembles those of foveaviruses.
Virion properties
Morphology
Virions are slightly flexuous filaments, 960 nm in length and 12–15 nm in diameter (Figure 5).
Physicochemical and physical properties
No information.
Nucleic acid
Virions contain a single molecule of positive sense ssRNA, about 8.7 kb long. There is a methylated cap at the 5’ terminus and a polyadenylated 3’ terminus.
Proteins
The viral capsid is composed of a single polypeptide of about 41 kDa.
Genome organization and replication
The genome contains three ORFs (Figure 6). ORFs 2 and 3 are separated by a short intergenic region and ORFs 1 and 2 overlap by 1 nt. ORF1 is the replication protein and is probably directly expressed from genomic RNA. It is assumed that the two smaller downstream ORFs, which code respectively for the putative “30K” MP and CP, are expressed via sgRNAs.
Antigenic properties
No information.
Biological properties
Host range
The virus causes abnormal bud union and leaf blotching in various citrus varieties. Citrus is the only known natural host and mechanical transmission to a range of herbaceous hosts has been unsuccessful.
Transmission
The virus is transmitted to citrus by grafting. There is no known natural vector.
Geographical distribution
The virus has been reported from citrus germplasm worldwide.
Cytopathic effects
No information.
Species demarcation criteria in the genus
Not applicable.
List of species in the genus Citrivirus
| Citrus leaf blotch virus |
|
|
| (Dweet mottle virus) |
|
|
| Citrus leaf blotch virus-SRA-153 | [AJ318061=NC_003877] | (CLBV-SRA153) |
Species names are in italic script; names of isolates are in roman script; names of synonyms are in roman script and parentheses. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Citrivirus but have not been approved as species
None reported.
Genus Foveavirus
Type species Apple stem pitting virus
Distinguishing features
Foveaviruses are distinct in having five ORFs and larger CPs than most other members of the family.
Virion properties
Morphology
Virions are flexuous filaments, about 800 to over 1000 nm in length and 12–15 nm in diameter with helical symmetry exhibiting a surface pattern with cross-banding and longitudinal lines (Figure 7). Particles of some viruses, including apple stem pitting virus (ASPV), show a tendency for end-to-end aggregation.
Physicochemical and physical properties
ASPV virions sediment as two or three bands in sucrose density gradients but yield a single band at equilibrium in Omnipaque 350 density gradients. They resist moderately high temperatures (thermal inactivation is around 60 °C) but not organic solvents, and are unstable in cesium chloride and sulfate.
Nucleic acid
Virions contain a single molecule of positive sense ssRNA, polyadenylated at the 3’ terminus.
Proteins
The viral capsid of all species is composed of a single polypeptide with a size ranging from 28 kDa (grapevine rupestris stem pitting-associated virus, GRSPaV) to 44 kDa (ASPV and apricot latent virus, ApLV).
Genome organization and replication
The genomes of all fully sequenced members contain five ORFs (Figure 8). The 5’ region initiates with a UTR of 33–72 nt, ORF1 codes for the replication-related protein, ORF2, ORF3 and ORF4 constitute the TGB and ORF5 is the CP cistron. A non-coding sequence of 176–312 nt followed by a poly(A) tail terminate the genome. ASPV virions accumulate in the cytoplasm, where multiplication is likely to occur following a strategy comparable to that of other viruses in the family, based on direct expression of the 5’-proximal ORF, and expression of downstream ORFs from sgRNAs. Multiple dsRNAs are found in infected hosts.
Antigenic properties
Antisera to ASPV that can be used for serological detection tests have been raised from purified virions or chimeric fusion CPs expressed in E. coli. ASPV and ApLV are serologically related, but there are no recognized serological relationships among other members of the genus.
Biological properties
Host range
The natural host range of individual species is restricted to a single (GRSPaV) or a few hosts (ASPV, ApLV). ASPV infects primarily pome fruits, causing diseases of apple (topworking disease) when grafted on susceptible rootstocks, of pear (vein yellows and necrotic spot) and quince. ApLV is the putative agent of peach asteroid spot and peach sooty ringspot diseases. GRSPaV is a pathogen of grapevine. Experimental host ranges are also restricted.
Transmission
No vector is known for any of the viruses. ASPV is transmitted by grafting and persists in the host propagative material. ASPV is mechanically transmissible, with some difficulty, to Nicotiana occidentalis and its subspecies obliqua.
Geographical distribution
All members have a wide geographical distribution.
Cytopathic effects
ASPV elicits a severe derangement of the cytology of infected cells but no specific cytopathic structures or inclusion bodies. Virus particles accumulate in bundles in the cytoplasm.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Natural host range.
- Serological specificity.
- CP size.
- Less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
List of species in the genus Foveavirus
| Apple stem pitting virus |
|
|
| Apple stem pitting virus-PA66 | [D21829=NC_003462] | (ASPV-PA66) |
| Apricot latent virus |
|
|
| Apricot latent virus-Caserta12 | [AF318062*] | (ApLV-Caserta12) |
| Grapevine rupestris stem pitting-associated virus |
|
|
| Grapevine rupestris stem pitting-associated virus-USA | [AF057136=NC_001948] | (GRSPaV-USA ) |
| Peach chlorotic mottle virus |
|
|
| Peach chlorotic mottle virus-Agua-4N6 | [EF693898=NC_009892] | (PCMoV-Agua4N6 ) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Foveavirus but have not been approved as species
| Asian prunus virus 1 | [FJ824737] | (APV1) |
| Asian prunus virus 2 | [DQ205237*] | (APV2) |
| Asian prunus virus 3 | [DQ205238*] | (APV3) |
* Sequences do not comprise the complete genome.
Genus Tepovirus
(Established 2011)
Type species: Potato virus T
List of species in the genus Tepovirus
| Potato virus T |
|
|
| Potato virus T-Peru | [EU835937=NC_011062] | (PVT-Peru) |
Genus Trichovirus
Type species Apple chlorotic leaf spot virus
Distinguishing features
Trichoviruses have three (or sometimes four) ORFs including a movement protein of the “30K” superfamily.
Virion properties
Morphology
Virions are very flexuous filaments, 640–890×10–12 nm in size, helically constructed with a pitch of 3.3–3.5 nm, and about 10 subunits per turn of the helix. Virions may show cross banding, criss-cross or rope-like features according to the negative contrast material used (Figure 9).
Physicochemical and physical properties
Virions sediment as single or as two very close bands with an S20,w of about 100S. Apple chlorotic leaf spot virus (ACLSV) virions are sensitive to ribonucleases. Virions of all viruses in the genus resist moderately high temperatures (thermal inactivation is around 55–60 °C) and are moderately resistant to organic solvents.
Nucleic acid
Virions contain a single molecule of linear, positive sense, ssRNA about 7.5– 8.0 kb in size, with a polyadenylated 3’ terminus, accounting for about 5% of the particle weight. Indirect evidence suggests that the genome RNA of ACLSV is capped at its 5’-end with m7G. An infectious cDNA clone of ACLSV has been produced. ACLSV isolates show a high variability in their nt sequence with an overall identity between 76 and 82%. The CP is the most conserved protein (87–93% identity), whilst the putative MP is the most divergent (77–85% identity).
Proteins
Virions of all members are composed of a single 20.5–27 kDa polypeptide.
Genome organization and replication
The genomes of ACLSV and grapevine berry inner necrosis virus (GINV) contain three slightly overlapping ORFs while other members and possible members of the genus have an additional ORF at the 3’ terminus (Figure 10). The large 5’ ORF of ACLSV is directly expressed from genomic RNA, whereas the two smaller downstream ORFs that code, respectively, for the MP and CP, are expressed via sgRNAs. The fourth ORF (where present) has homologies to the vitivirus nucleic acid binding proteins. ACLSV-infected tissues contain six dsRNA species of approximately 7.5, 6.4, 5.4, 2.2, 1.1 and 1.0 kbp. The 7.5 kbp species represents the double-stranded form of the full-length genome, whereas the 2.2 and the 1.1 kbp species are the double-stranded forms of sgRNAs coding for the MP and the CP, respectively. The most abundant dsRNA species, the function of which are unknown, are 5’ co-terminal with genomic RNA, and have sizes of 6.4 and 5.4 kbp, respectively. Replication is presumed to be cytoplasmic and to involve the translation product of ORF1. The MP of ACLSV is a suppressor of silencing that interferes with systemic movement of the silencing signal.
Antigenic properties
Virions are moderate to poor antigens. Cherry mottle leaf virus (CMLV) and peach mosaic virus (PcMV) are serologically related to one another but not to the other members of the genus.
Biological properties
Host range
The natural host range of individual species is relatively narrow (ACLSV, PcMV), or restricted to a single host (GINV, CMLV). The experimental host range is somewhat wider, but still limited to a few herbaceous species. In the natural hosts, infections induce few or no symptoms (ACLSV in certain hosts), or mottling, rings, line patterns and fruit injuries (i.e. pseudosharka) (ACLSV), mottling with stunting and internal necrosis of shoots and berries (GINV), mottling and severe distortion of the leaves (CMLV), mottling and deformation of leaves and fruits and color break in the petals (PcMV).
Transmission
The viruses are readily transmitted by mechanical inoculation, by grafting (ACLSV, GINV, CMLV, PcMV) and through propagating material. GINV is transmitted by the grape erineum mite Colomerus vitis, CMLV by the scale mite Eriophyes inequalis, and PcMV by the peach bud mite Eriophyes insidiosus.
Geographical distribution
Geographical distribution varies from wide to restricted, according to the virus. ACLSV is ubiquitous, whereas GINV is reported only from Japan, and CMLV and PcMV from North America.
Cytopathic effects
Infected cells are damaged by ACLSV to varying extents. Virions are found in phloem and parenchyma cells of leaves and roots and accumulate in the cytoplasm, sometimes in the nucleus, in bundles or paracrystalline aggregates. No inclusion bodies are formed.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- Natural and experimental host range.
- Serological specificity.
- Less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
- Transmission by a vector.
- Vector specificity.
List of species in the genus Trichovirus
| Apple chlorotic leaf spot virus |
|
|
| Apple chlorotic leaf spot virus-P863 | [M58152=NC_001409] | (ACLSV-P863) |
| Apricot pseudo-chlorotic leaf spot virus |
|
|
| Apricot pseudo-chlorotic leaf spot virus-Sus2 | [AY713379=NC_006946] | (APsCLSV-Sus2) |
| Cherry mottle leaf virus |
|
|
| Cherry mottle leaf virus-SA1162-21 | [AF170028=NC_002500] | (CMLV-SA116221) |
| Grapevine berry inner necrosis virus |
|
|
| Grapevine berry inner necrosis virus-Japan | [D88448=NC_015220] | (GINV-JP) |
| Peach mosaic virus |
|
|
| Peach mosaic virus-2022-01 (CA-1) | [DQ117579=NC_011552] | (PcMV-202201 (CA1)) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the genus Trichovirus but have not been approved as species
| Fig latent virus 1 | [FN377573*] | (FLV-1) |
| Phlomis mottle virus | [AM920542*] | (PhMV) |
* Sequences do not comprise the complete genome.
Genus Vitivirus
Type species Grapevine virus A
Distinguishing features
Vitiviruses have a distinctive genome organization with five ORFs, including a 20K ORF between the polymerase and the movement protein of the “30K” superfamily. Natural transmission is by pseudococcid mealybugs, soft scale insects and aphids.
Virion properties
Morphology
Virions are flexuous filaments 725–825×12 nm in size, showing distinct cross-banding, helically constructed with a pitch of 3.3–3.5 nm and about 10 subunits per turn of the helix (Figure 11).
Physicochemical and physical properties
Virions sediment as a single or two very close bands in sucrose or Cs2SO4 gradients, with an S20,w of about 92S. Virions of Heracleum latent virus (HLV) are sensitive to ribonucleases. Virions of all members of the genus resist moderately high temperatures (thermal inactivation is around 60 °C) and are moderately resistant to organic solvents.
Nucleic acid
Virions contain a single molecule of positive sense ssRNA, about 7.6 kb in size, capped at the 5’ terminus with m7G and polyadenylated at the 3’ terminus. The RNA accounts for about 5% of the particle weight. Infectious cDNA clones have been produced for grapevine viruses A and B (GVA and GVB).
Proteins
The CPs are composed of a single 18–21.5 kDa polypeptide.
Genome organization and replication
The genomes contain five slightly overlapping ORFs (Figure 12). The 5’ regions of grapevine viruses A and B initiate with an A/T-rich (60–68%) UTR of 47–86 nt. ORF1 is the replication-related protein. ORF2 is a 19–20 kDa polypeptide of unknown function with no significant sequence homology to known proteins, which, in GVB infections, does not accumulate in phase with MPs. ORF3 (31–36.5 kDa) is the movement protein and ORF4 is the CP. The final ORF is a 10–14 kDa polypeptide with weak homologies to proteins with RNA-binding properties and which has been shown (in GVA) to have RNA silencing suppressor activity.
The strategy of expression is based on sgRNA production, as suggested by the analysis of dsRNA patterns from infected tissues. The four dsRNAs have sizes of 7.6, 6.48, 5.68 and 5.1 kbp for GVA and GVD, and 7.6, 6.25, 5.03 and 1.97 kbp for GVB. In GVA there are nested sets of 5’-terminal sgRNAs 5.1, 5.5 and 6.0 kb in size and of 3’-terminal sgRNAs 1.0, 1.8 and 2.2 kb that serve for the expression of all ORFs, except for ORF5, which may be expressed via a bi- or polycistronic mRNA. The generation of these 5’- and 3’-terminal sgRNAs appears to be controlled by internal cis-acting elements. Replication occurs in the cytoplasm, possibly in association with membranous vesicles.
Antigenic properties
Virions are moderate or poor antigens. Most species are very distantly serologically related. Monoclonal antibodies to GVA, GVB and GVD and recombinant protein antibodies to the putative MP of GVA have been produced. The relationship between GVA, GVB and GVD is due to a few common internal antigenic determinants (cryptotopes). GVA particles carry a highly structured epitope centered in a common peptide region of the CP sequence.
Biological properties
Host range
The natural host range of individual species is restricted to a single host. Infections induce either no symptoms (HLV and mint virus 2 (MV-2)) or severe diseases characterized by pitting and grooving of the wood (GVA, GVB and GVD). The experimental host range varies from wide (HLV) to restricted (GVA, GVB, GVD and GVE).
Transmission
All species except for MV-2 are transmitted by mechanical inoculation, those infecting grapevines with greater difficulty. Transmission by grafting and dispersal through propagating material is common with grapevine-infecting species. GVA and GVB are transmitted in a semi-persistent manner by different species of pseudococcid mealybugs of the genera Pseudococcus and Planococcus. GVA is also transmitted by the scale insect Neopulvinaria innumerabilis. HLV and MV-2 are transmitted semi-persistently by aphids, in association with a helper virus.
Geographical distribution
Geographical distribution varies from very wide (GVA, GVB, GVD) to restricted (HLV).
Cytopathic effects
Infected cells are damaged to a varying extent. All species investigated elicit the formation of vesicular evaginations of the tonoplast containing finely fibrillar material, possibly representing replicative forms of viral RNA. Virions of grapevine-infecting species are strictly phloem-limited, but in herbaceous hosts they also invade the parenchyma. Virus particles accumulate in the cytoplasm in bundles or paracrystalline aggregates.
Species demarcation criteria in the genus
The criteria demarcating species in the genus are:
- The natural host range.
- Serological specificity using discriminatory polyclonal and monoclonal antibodies.
- Epidemiology: individual species or groups of species are transmitted by different types and species of vectors.
- Differences in dsRNA pattern.
- Less than about 72% nt identity (or 80% aa identity) between their CP or polymerase genes.
List of species in the genus Vitivirus
| Grapevine virus A |
|
|
| Grapevine virus A-Is 151 | [X75433=NC_003604] | (GVA-Is 151) |
| Grapevine virus B |
|
|
| Grapevine virus B-Italy | [X75448=NC_003602] | (GVB-IT) |
| Grapevine virus D |
|
|
| Grapevine virus D-Italy | [Y07764*] | (GVD-IT) |
| Grapevine virus E |
|
|
| Grapevine virus E-Japan:TvAQ7 | [AB432910=NC_011106] | (GVE-TvAQ7) |
| Heracleum latent virus |
|
|
| Heracleum latent virus-Scottish: Murant | [X79270*] | (HLV-MUR) |
| Mint virus 2 |
|
|
| Mint virus 2-USA | [AY913795*] | (MV-2-USA) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
* Sequences do not comprise the complete genome.
List of other related viruses which may be members of the genus Vitivirus but have not been approved as species
None reported.
List of unassigned species in the family Betaflexiviridae
| African oil palm ringspot virus |
|
|
| African oil palm ringspot virus-Colombia | [AY072921=NC_012519] | (AOPRV-COL) |
| Banana mild mosaic virus |
|
|
| Banana mild mosaic virus-Australia | [AF314662=NC_002729] | (BanMMV-AUS) |
| Cherry green ring mottle virus |
|
|
| Cherry green ring mottle virus-USA | [AF017780=NC_001946] | (CGRMV-USA) |
| Cherry necrotic rusty mottle virus |
|
|
| Cherry necrotic rusty mottle virus-Germany | [AF237816=NC_002468] | (CNRMV-DE) |
| Sugarcane striate mosaic-associated virus |
|
|
| Sugarcane striate mosaic-associated virus-Australia | [AF315308=NC_003870] | (SCSMaV-AUS) |
Species names are in italic script; names of isolates are in roman script. Sequence accession numbers [ ] and assigned abbreviations ( ) are also listed.
List of other related viruses which may be members of the family Betaflexiviridae but have not been approved as species
| Banana virus X | [AY710267*] | (BanVX) |
| White ash mosaic virus | [DQ412998= NC_011533] | (WAMV) |
* Sequences do not comprise the complete genome.
Phylogenetic relationships within the family
In a phylogenetic analysis of the replication protein, most genera fall on well-supported branches (Figure 13, p. 938). The family falls into two broad parts that correspond with the types of movement protein. Carlavirus and Foveavirus with a number of unassigned species that have a TGB form one branch, while the remaining genera and viruses (which all have a “30K”-type movement protein) also cluster together. Of the unassigned species, Potato virus T is expected to become the type member of a new genus (proposed Tepovirus). The remaining unassigned species resemble foveaviruses in their genome organization but do not form a monophyletic group.
Similarity with other taxa
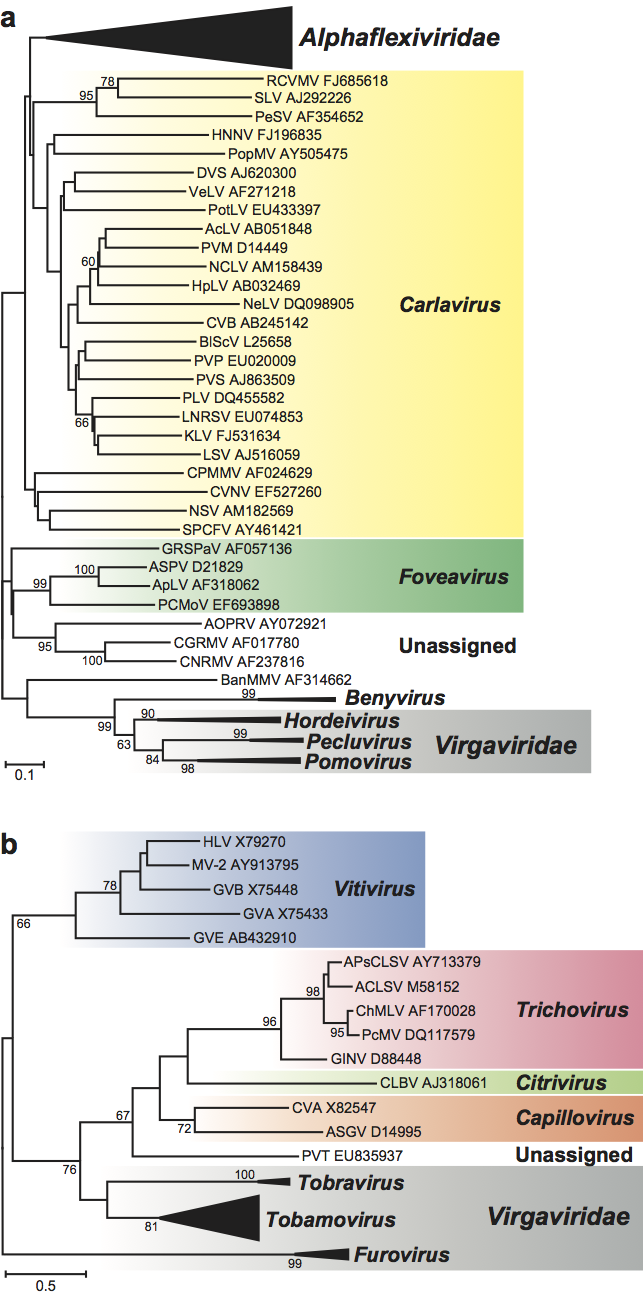
The polymerase proteins are members of the “alphavirus-like” supergroup of RNA viruses and are most closely related to those of the other families in the order, namely Alphaflexiviridae, Gammaflexiviridae and Tymoviridae. There are also similarities between the TGB-containing genera and members of the family Alphaflexviridae in the 3’ end of the genome (Figure 14, p. 939). The TGB proteins are also more distantly related to those of rod-shaped viruses in the family Virgaviridae (genera Hordeivirus, Pecluvirus and Pomovirus) (Figure 15a, p. 940). The “30K” movement proteins are related to those in the family Virgaviridae (genera Furovirus, Tobamovirus and Tobravirus) (Figure 15b, p. 940).
Derivation of names
Capillo: from Latin capillus, “a hair”.
Carla: from Carnation latent virus.
Citri: from Citrus leaf blotch virus.
Flexi: from Latin flexus, “bent”.
Fovea: from Latin fovea, “pit” or “hole”, a type of symptom induced by the type species.
Tricho: from Greek thrix, “hair”.
Viti: from Vitis, generic name of the grapevine, Vitis vinifera, the host of the type species.
Further reading
Journals and books
Adams et al., 2004 M.J. Adams, J.F. Antoniw, M. Bar-Joseph, A.A. Brunt, T. Candresse, G.D. Foster, G.P. Martelli, R.G. Milne, S.K. Zavriev, C.M. Fauquet, The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch. Virol. 149 (2004) 1045–1060.
Bratlie and Drabløs, 2005 M.S. Bratlie, F. Drabløs, Bioinformatic mapping of AlkB homology domains in viruses. BMC Genomics. 6 (2005) 1–15.
Martelli et al., 2007 G. Martelli, M.J. Adams, J.F. Kreuze, V.V. Dolja, Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45 (2007) 73–100.
Vives et al., 2001 M.C. Vives, L. Galipienso, L. Navarro, P. Moreno, J. Guerri, The nucleotide sequence and genomic organization of citrus leaf blotch virus: candidate type species for a new virus genus. Virology. 287 (2001) 225–233.
Websites
International Council for the Study of Virus and Virus-like Diseases of the Grapevine: http://www.icvg.ch.
Contributed by
Adams, M.J., Candresse, T., Hammond, J., Kreuze, J.F., Martelli, G.P., Namba, S., Pearson, M.N., Ryu, K.H., Saldarelli, P. and Yoshikawa, N.
Figures
Figure 1 (Left) Schematic representation of a fragment of a particle of a capillovirus. (Right) Negative contrast electron micrograph of particles of an isolate of the species Apple stem grooving virus. The bar represents 100 nm.
(Courtesy of N. Yoshikawa.)
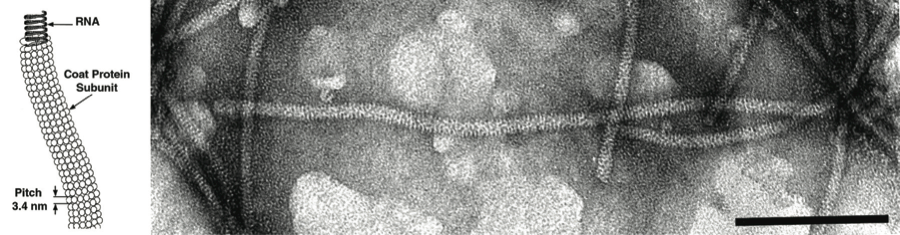
Figure 2 Genome organization of apple stem grooving virus showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; P-Pro, papain-like protease; Hel, helicase; RdRp, RNA-dependent RNA polymerase; MP, putative movement protein; CP, capsid protein.

Figure 3 Filamentous particles of an isolate of the species Carnation latent virus. The bar represents 100 nm.
(Courtesy R.G. Milne.)
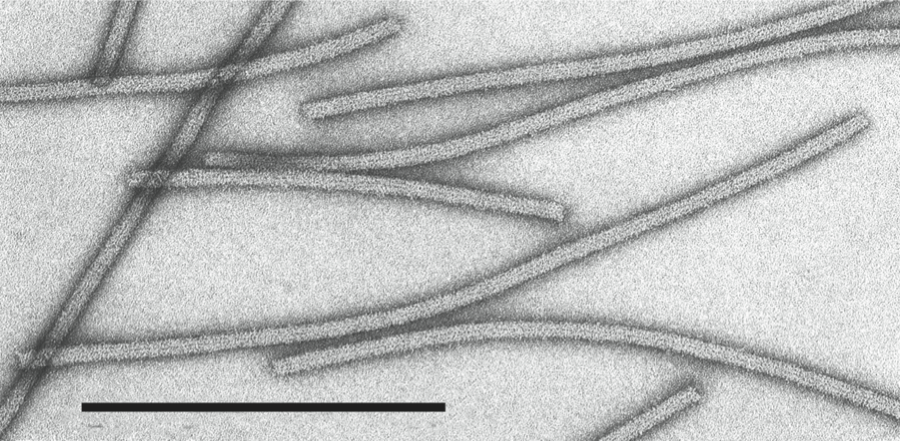
Figure 4 Genome organization of potato virus M showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; P-Pro, papain-like protease; Hel, helicase; RdRp, RNA-dependent RNA polymerase; CP, capsid protein; NB, nucleic acid binding protein. The 25K, 12K and 7K proteins constitute the triple gene block.

Figure 5 Negative contrast electron micrograph of particles of an isolate of the species Citrus leaf blotch virus. The bar represents 200 nm.
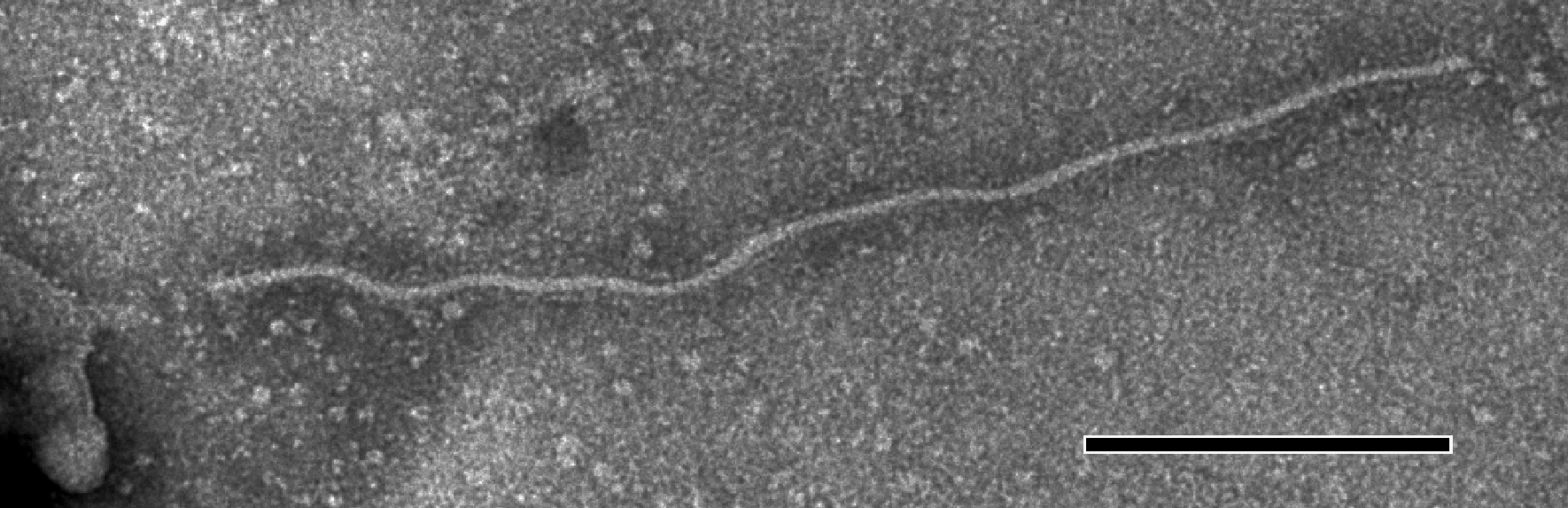
Figure 6 Genome organization of citrus leaf blotch virus showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; P-Pro, papain-like protease; Hel, helicase; RdRp, RNA-dependent RNA polymerase; MP, putative movement protein; CP, capsid protein.
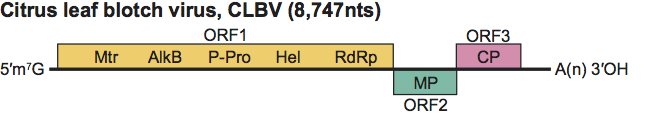
Figure 7 Negative contrast electron micrograph of particles of an isolate of the species Apple stem pitting virus. The bar represents 100 nm.
(Courtesy of H. Koganezawa.)
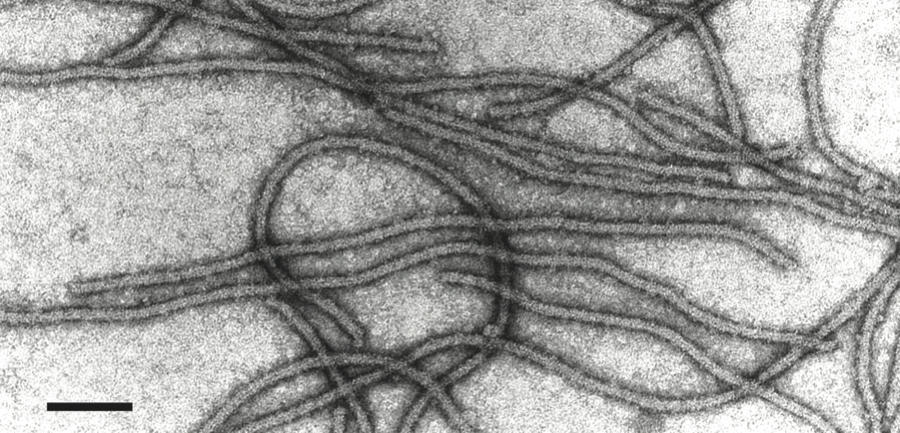
Figure 8 Genome organization of apple stem pitting virus showing the relative position of the ORFs and their expression products. Mtr, methytransferase; P-Pro, papain-like protease; Hel, helicase; Pol, polymerase; TGB, triple gene block; CP, capsid protein.
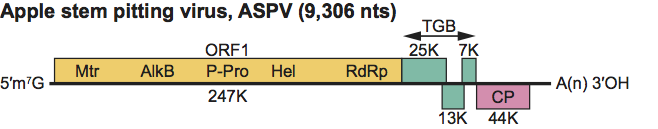
Figure 9 Negative contrast electron micrograph of particles of an isolate of the species Apple chlorotic leaf spot virus. The bar represents 100 nm.
(Courtesy of M.A. Castellano.)
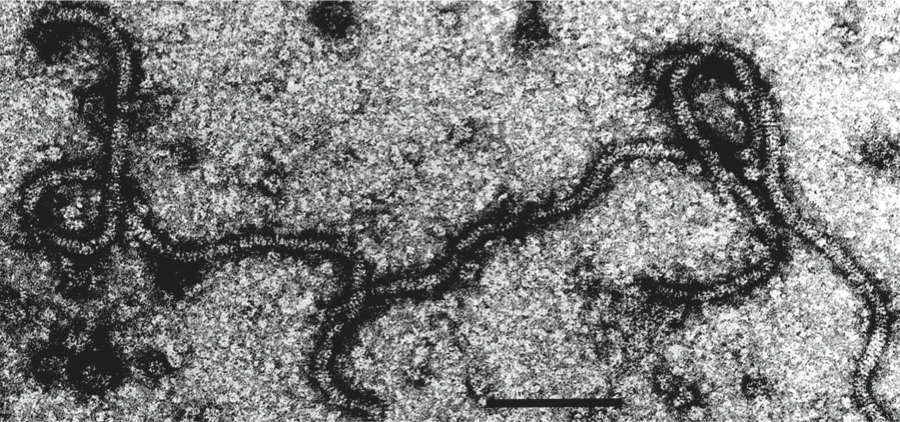
Figure 10 Genome organization of apple chlorotic leaf spot virus and cherry mottle leaf virus, showing the relative positions of the ORFs and their expression products. Mtr, methyltransferase; P-Pro, papain-like protease; Hel, helicase; Pol, polymerase; MP, putative movement protein; CP, capsid protein; NB, nucleic acid binding protein.
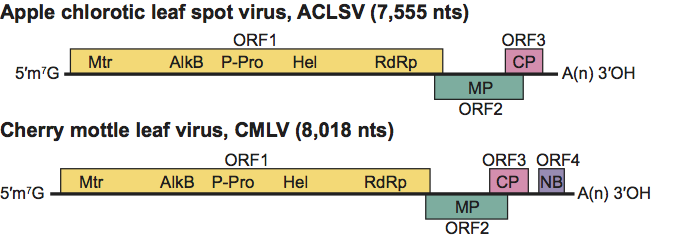
Figure 11 Negative contrast electron micrograph of particles of an isolate of the species Grapevine virus A. The bar represents 100 nm.
(Courtesy of A.A. Castellano.)
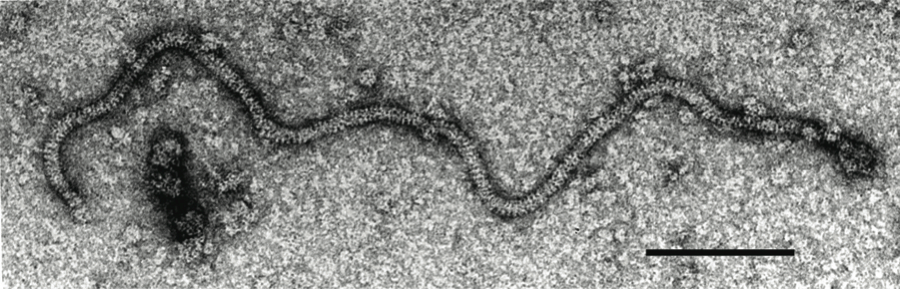
Figure 12 Organization and expression of the genome of grapevine virus A (GVA) showing the relative position of the ORFs, their expression products, and the nested sets of 5- and 3-terminal sgRNAs. Mtr, methyltransferase; Hel, helicase; Pol, polymerase; MP, putative movement protein; CP, capsid protein.
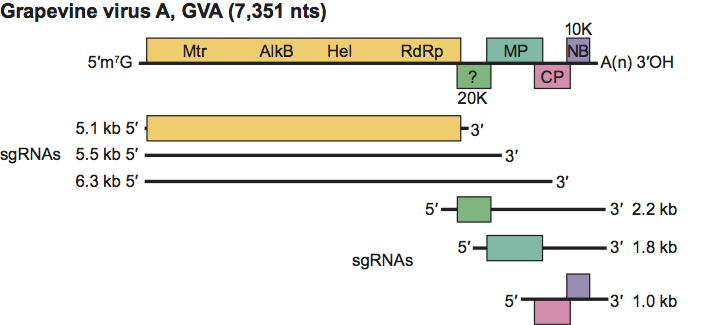
Figure 13 Phylogenetic (distance) tree based on the amino acid sequences of the entire replication protein of members of the family Betaflexiviridae. A single representative isolate of each sequenced species in the family was included and the tree is rooted with Botrytis virus F (BotV-F; genus Mycoflexivirus, family Gammaflexiviridae). Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances. Tree produced in MEGA4.
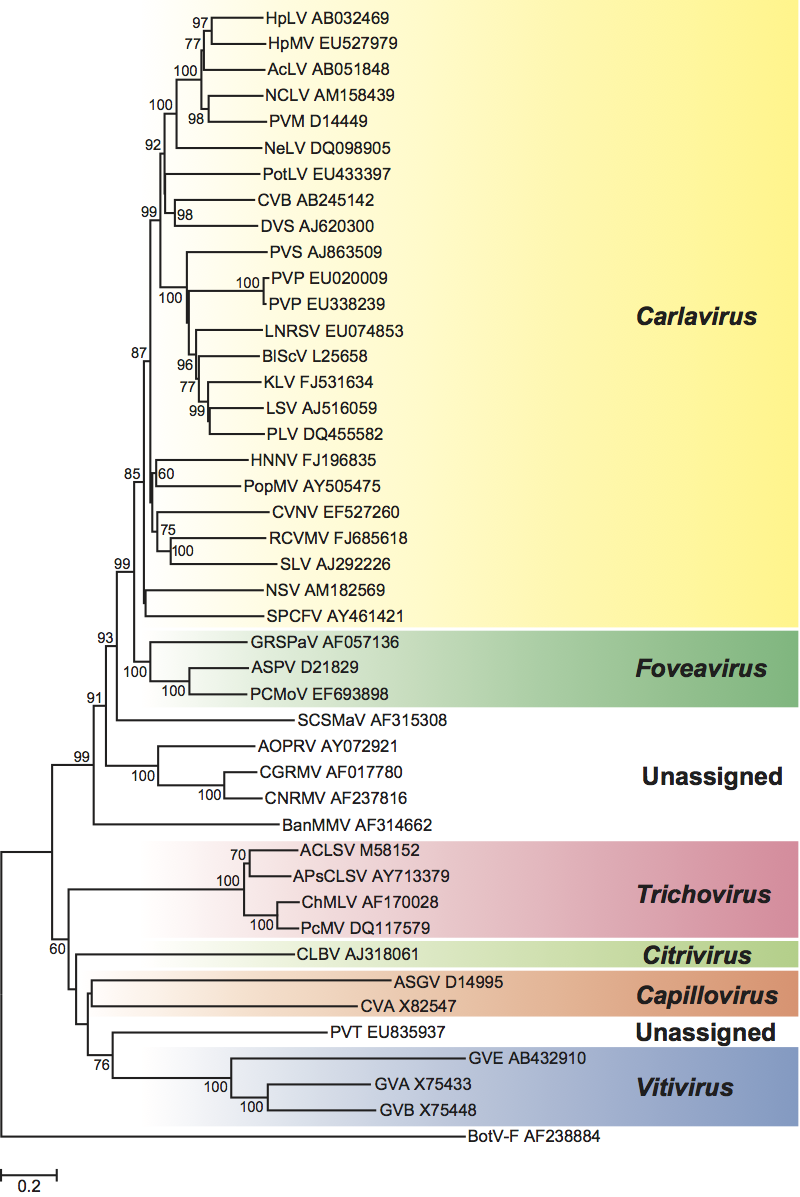
Figure 14 Phylogenetic (distance) tree based on the 3-terminal nucleotide sequences of those members of the families Betaflexiviridae and Alphaflexiviridae that have a triple gene block (TGB). The sequence used includes the entire TGB and coat protein genes (alignment length 2466 nt). A single representative isolate of each sequenced species in each family was used. Numbers on branches indicate percentage of bootstrap support out of 10,000 bootstrap replications (when >60%). The scale indicates maximum composite likelihood distances. Tree produced in MEGA4.
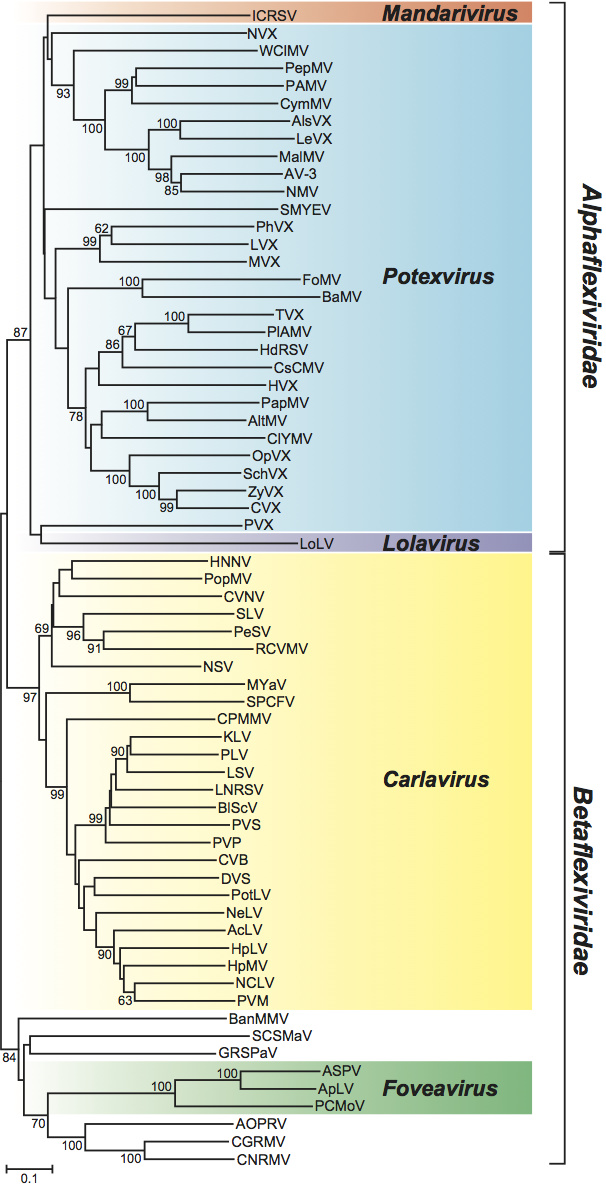
Figure 15 Phylogenetic (distance) trees produced in MEGA4 showing how the cell-to-cell movement proteins of viruses in the family Betaflexiviridae are related to those of other plant viruses. (a) Tree based on the codon-aligned nucleotide sequences of the first triple gene block protein (TGBp1). A single representative isolate of each sequenced species in the family was included. The tree also contains similar sequences from the family Alphaflexiviridae and those of rod-shaped plant viruses collapsed into triangles, the length of which corresponds to the variation found within the clade. Numbers on branches indicate percentage of bootstrap support out of 10,000 bootstrap replications (when >60%). The scale indicates maximum composite likelihood distances. Tree produced in MEGA4. (b) Tree based on the amino acid sequences of the 30K-like movement protein. A single representative isolate of each sequenced species in the family was included. The tree also contains similar sequences from genera in the family Virgaviridae collapsed into triangles, the length of which corresponds to the variation found within the clade. Numbers on branches indicate percentage of bootstrap support out of 1000 bootstrap replications (when >60%). The scale indicates JTT amino acid distances.