Family: Filoviridae
Nadine Biedenkopf, Alexander Bukreyev, Kartik Chandran, Nicholas Di Paola, Pierre B. H. Formenty, Anthony Griffiths, Adam J. Hume, Elke Mühlberger, Sergey V. Netesov (Нетёсов Сергей Викторович), Gustavo Palacios, Janusz T. Pawęska, Sophie Smither, Ayato Takada (高田礼人), Victoria Wahl and Jens H. Kuhn
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Jens H. Kuhn (kuhnjens@mail.nih.gov)
Edited by: Jens H. Kuhn, Stuart G. Siddell, and Peter J. Walker
Posted: March 2019, updated October 2020, January 2024, June 2024
Summary
Filoviridae is a family for negative-sense RNA viruses with genomes of about 13.1–20.9 kb that infect fish, mammals, and reptiles (Table 1.Filoviridae). The filovirid genome is a linear, non-segmented RNA with 5 canonical open reading frames (ORFs) that encode a nucleoprotein (NP), a polymerase cofactor (VP35), a glycoprotein (GP1,2), a transcriptional activator (VP30), and a large protein (L) containing an RNA-directed RNA polymerase (RdRP) domain. All filovirids genomes encode additional proteins that vary among genera. Several filovirids (e.g., Ebola virus, Marburg virus) are pathogenic for humans and are highly virulent.
Table 1.Filoviridae. Characteristics of members of the family Filoviridae.
| Characteristic | Description |
| Example | Marburg virus (DQ217792), species Orthomarburgvirus marburgense |
| Virion | Enveloped, variously shaped but predominantly filamentous, typically with a single nucleocapsid |
| Genome | 13.1–20.9 kb of linear, negative-sense, non-segmented RNA |
| Replication | The genome forms ribonucleoprotein complexes, which serve as templates for transcription and replication. Encapsidated antigenomic RNA is a replication intermediate |
| Translation | From multiple monocistronic 5′-capped and 3′-polyadenylated mRNAs |
| Host range | Fish (oblaviruses, striaviruses, thamnoviruses), mammals (cuevaviruses, dianloviruses, orthoebolaviruses, orthomarburgviruses), reptiles (tapjoviruses) |
| Taxonomy | Realm Riboviria, phylum Negarnaviricota, subphylum Haploviricotina, class Monjiviricetes, order Mononegavirales; the family includes 9 genera and 17 species |
Filovirids form a monophyletic clade based on phylogenetic analysis of RNA-directed RNA polymerase (RdRP) sequences (Wolf et al., 2018). Genomes of viruses from all eight genera have a similar genomic architecture but differ in the number of open reading frames (ORFs) and the number and location of gene overlaps (Kuhn et al., 2020).
Piscine Hosts
Genus Oblavirus. This genus includes one species for one virus (Oberland virus [OBLV]), discovered in diseased farmed European perch (family Percidae) exported from Germany to Switzerland. Oblaviruses are notable for genomes that contain at least two proteins without obvious homologs in viruses of other filovirid genera, and do not encode matrix protein (VP40) or RNP complex associated protein (VP24) encoded by all mammalian filovirids (Hierweger et al., 2021). Oblaviruses have not been cultured to date.
Genus Striavirus. This genus includes one species for one virus (Xīlǎng virus [XILV]), discovered in captured frogfish (family Antennariidae) from the East China Sea. Striaviruses are notable for encoding at least four proteins without obvious homologs in other filovirid genera, and do not encode VP24 (Shi et al., 2018, Hume and Mühlberger 2019). Striaviruses have not been cultured to date.
Genus Thamnovirus. This genus includes three species for three viruses (Fiwi virus [FIWIV], Kander virus [KNDV], Huángjiāo virus [HUJV]), discovered in captured filefish (family Monacanthidae) from the East China Sea and in diseased farmed European perch (family Percidae) exported from Germany to Switzerland. Thamnoviruses are notable for genomes that encode at least two proteins without obvious homologs in viruses of other filovirid genera and do not encode matrix protein (VP40) or VP24 (Shi et al., 2018, Hume and Mühlberger 2019, Hierweger et al., 2021). Thamnoviruses have not been cultured to date.
Mammalian Hosts
Genus Cuevavirus. This genus includes one species for one virus (Lloviu virus [LLOV]), discovered in miniopterid bats in Europe. The organization of cuevavirus genomes is reminiscent of orthoebolavirus genomes. Similar to orthoebolaviruses, cuevaviruses encode VP24 and VP40, and GP1,2 production relies on RNA editing (Negredo et al., 2011, Kemenesi et al., 2018, Kemenesi et al., 2022, Tóth et al., 2023).
Genus Dianlovirus. This genus includes one species for one virus (Měnglà virus [MLAV]), discovered in pteropodid bats. Dianloviruses have only been reported from Asia (Yang et al., 2017, Yang et al., 2019, Paskey et al., 2020, Makenov et al., 2023). The organization of dianlovirus genomes is highly reminiscent of orthomarburgvirus genomes (Yang et al., 2019). Similar to orthomarburgviruses, dianloviruses encode VP24 and VP40, and RNA editing is not required for GP1,2 production. Dianloviruses have not been cultured to date.
Genus Orthoebolavirus. This genus includes six species for six viruses. One of these viruses, Bombali virus (BOMV), has been detected in molossid bats (Goldstein et al., 2018b). Infectious BOMV has been generated using recombinant DNA technology. No naturally occurring BOMV isolates have been cultured to date. Two additional viruses, Ebola virus (EBOV) and Reston virus (RESTV), are suspected to be harbored by bats as natural hosts. Five orthoebolaviruses (Bundibugyo virus [BDBV], EBOV, RESTV, Sudan virus [SUDV], and Taï Forest virus [TAFV]) are pathogenic for nonhuman primates. BDBV, EBOV, and SUDV are highly lethal human pathogens. TAFV has caused only a single case of severe but non-lethal human disease, and RESTV has only caused one inapparent human infection. RESTV has also been found in domestic pigs. RESTV appears to be endemic in South-eastern Asia; all other orthoebolaviruses circulate in Africa (Kuhn et al., 2020). Orthoebolaviruses encode VP40 and VP24. They are notable for expressing three distinct proteins from their glycoprotein (GP) genes, a strategy they share with cuevaviruses (Volchkov et al., 1995, Sanchez et al., 1996, Negredo et al., 2011).
Genus Orthomarburgvirus. This genus includes one species for two viruses found in pteropodid bats in Africa. Both viruses (Marburg virus [MARV] and Ravn virus [RAVV]) are highly lethal human pathogens (Kuhn et al., 2020). Orthomarburgviruses encode VP40 and VP24.
Reptilian Hosts
Genus Tapjovirus. This genus includes one species for one virus (Tapajós virus [TAPV]), discovered during a metagenomic analysis in a viperid snake from Brazil. Tapjoviruses are notable for genomes that are closely related to piscine filovirids (striaviruses) but having the genomic organization of mammalian filovirids (Horie 2021). Tapjoviruses have not been cultured to date.
Virion
Morphology
Virion morphology (Figure 1.Filoviridae) has only been studied for cuevaviruses, orthoebolaviruses, and orthomarburgviruses and is described in the respective genus subchapter pages.
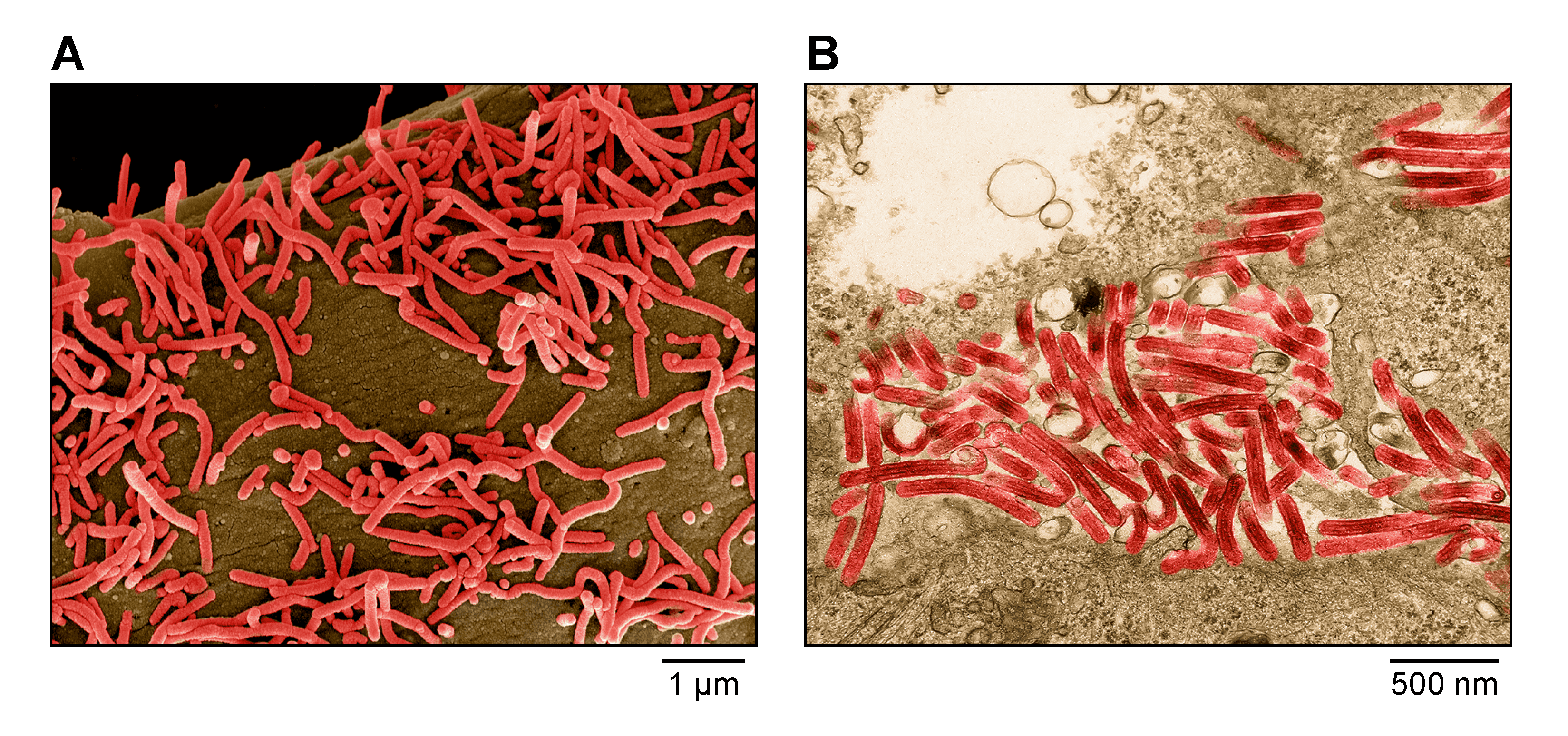 |
| Figure 1. Filoviridae. A) Scanning electron micrograph of Marburg virus particles (red) budding from an infected grivet (Chlorocebus aethiops (Linnaeus, 1758)) Vero E6 cell. B) Transmission electron micrograph of Marburg virus particles (red) found both as extracellular particles and budding particles from Vero E6 cells. Images are colorized for clarity. Courtesy of John G. Bernbaum and Jiro Wada, NIH/NIAID/DCR/IRF Frederick, Fort Detrick, MD, USA. |
Physicochemical and physical properties
Physicochemical and physical properties have only been described for individual orthoebolaviruses and orthomarburgviruses and are described in the respective genus pages.
Nucleic acid
Filovirid genomes are linear non-segmented RNA molecules of negative polarity. The genomes vary from about 13.1 kb (thamnoviruses) to about 19 kb (cuevaviruses, orthoebolaviruses, and orthomarburgviruses) (Feldmann et al., 1992, Sanchez et al., 1993, Negredo et al., 2011, Shi et al., 2018). The genome of an unclassified virus from pteropodid bats, Déhóng virus (DEHV), reaches 20.9 kb (He et al., 2023).
Proteins
Filovirids express 6–10 proteins depending on genus. RNP complexes are composed of a genomic RNA molecule and several structural proteins, including nucleoprotein, VP35, VP30, and L (Hume and Mühlberger 2019).
Lipids
The filovirion envelope is derived from host cell membranes and is considered to have a lipid composition similar to that of the host-cell plasma membrane (Bavari et al., 2002). Some filovirid proteins may be acylated (Funke et al., 1995, Ito et al., 2001).
Carbohydrates
Carbohydrate composition has only been described for individual orthoebolaviruses and orthomarburgviruses and is described in the respective genus pages.
Genome organization and replication
Filovirid genomes are organized like most mononegaviral genomes, with the general mononegaviral gene order 3′ N P M (G) L 5′ (terminology for filovirids: 3′-NP-VP35-(VP40)-(GP)-L-5′), but differ in that they may contain additional genes (Figure 2.Filoviridae) (Feldmann et al., 1992, Sanchez et al., 1993, Negredo et al., 2011, Shi et al., 2018, Yang et al., 2019, Hierweger et al., 2021, Horie 2021, Seuberlich et al., 2023). The extragenic sequences at the extreme 3′-end (leader) and 5′-end (trailer) of filovirid genomes are conserved, and short sections of these end sequences are complementary. Genes of non-fish filovirids are flanked by conserved transcriptional initiation and termination (polyadenylation) sites typically containing the highly conserved pentamer 3′-UAAUU-5′. While containing divergent transcriptional initiation sites, striaviruses and thamnoviruses retain relatively conserved transcriptional termination sites(Hume and Mühlberger 2019). Genes may be separated by non-conserved intergenic sequences or overlaps. Most genes possess relatively long 3′- and 5′-noncoding regions. Co-transcriptional editing is used by cuevaviruses and orthoebolaviruses to express nonstructural proteins from the GP gene (Brauburger et al., 2015, Hume and Mühlberger 2019, Kuhn et al., 2020).
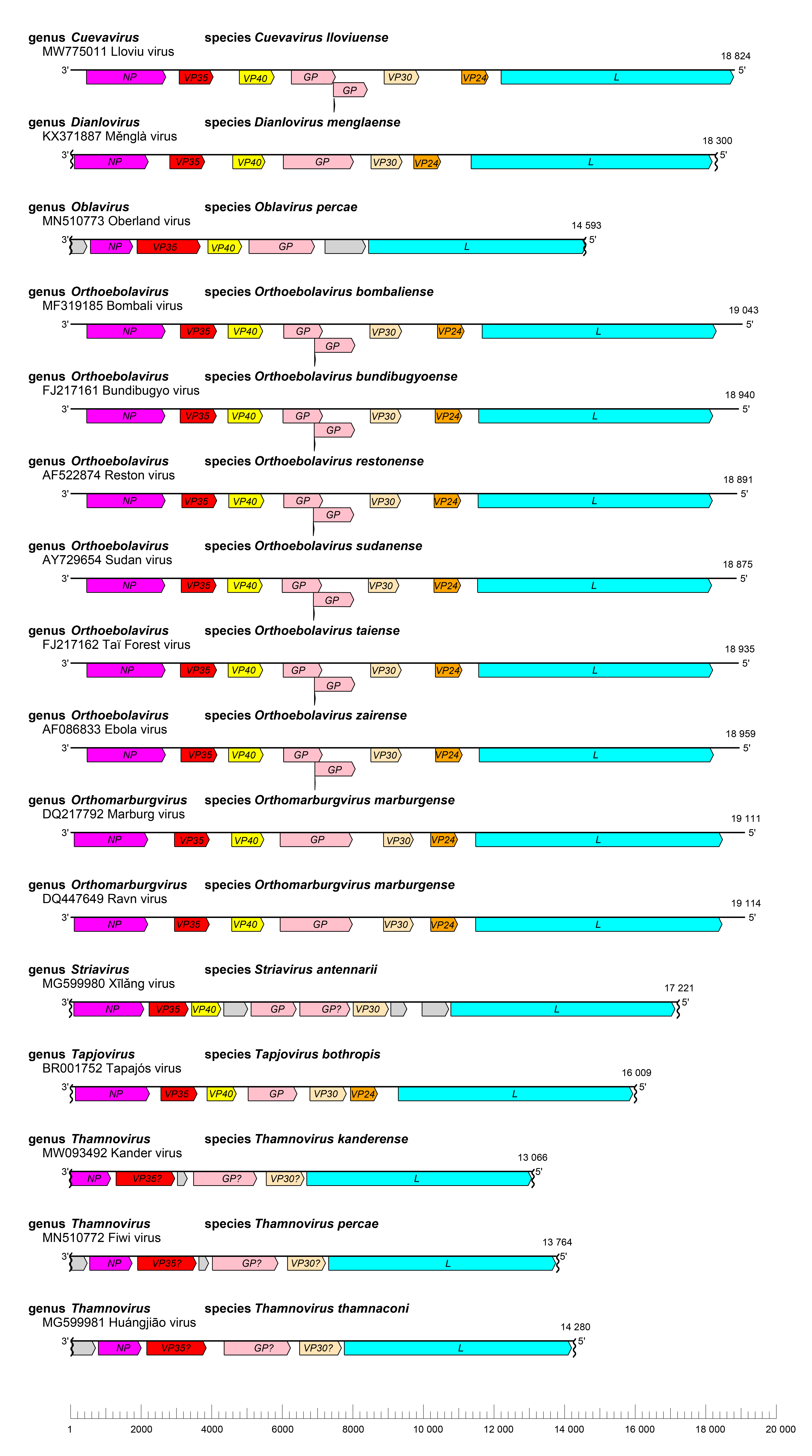 |
| Figure 2. Filoviridae. Schematic representation of filovirid genome organization. Genomes are drawn to scale. Wavy lines indicate incomplete genome ends. |
The replication strategies of filovirids (Figure 3.Filoviridae) have only been studied in depth using EBOV and MARV and are discussed in the respective Orthoebolavirus and Orthomarburgvirus genus subchapters.
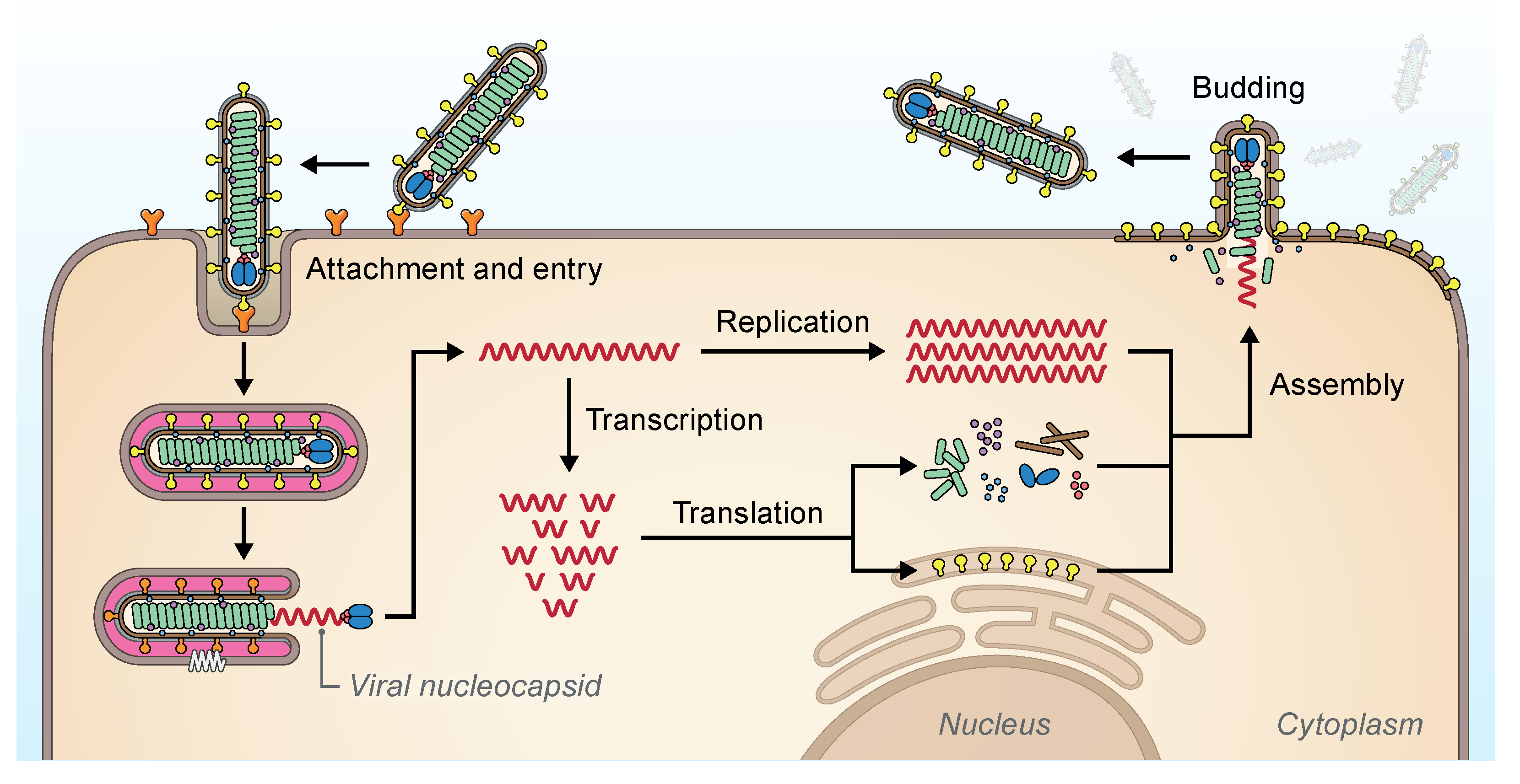 |
| Figure 3. Filoviridae. Replication cycle of filovirids (possibly excluding oblaviruses, striaviruses, and thamnoviruses). Virions attach to cell-surface attachment factors (orange Ys) and are taken into the cell via endocytosis (Davey et al., 2017). The filovirion glycoproteins (yellow clubs) bind to endosomal NPC intracellular cholesterol transporter 1 (NPC1, white zigzag) and catalyze the fusion of viral and cellular membranes to release the filovirid RNP complex (green helix) (Carette et al., 2011, Côté et al., 2011, Ng et al., 2014). The polymerase complex (consisting of VP35 [purple dots] and L [blue ovals]) transcribes filovirid mRNAs, which are translated into filovirid proteins, and replicates filovirid genomic RNA via antigenomic intermediates (Brauburger et al., 2015). Genomic RNA and antigenomic RNA occur only as ribonucleoprotein complexes, which serve as templates for replication and/or transcription. Assembly of filovirid proteins and progeny genomes occurs in the cytoplasm and results in budding and release of virions at the plasma membrane (Kolesnikova et al., 2017). Courtesy of Jiro Wada, NIH/NIAID/DCR/IRF-Frederick, Fort Detrick, MD, USA. |
Biology
Filovirids are endemic in Eastern Africa (BDBV, MARV, RAVV, SUDV), Middle Africa (BDBV, EBOV, MARV), Southern Africa (MARV), Western Africa (BOMV, EBOV, MARV, TAFV), South America (TAPV), Eastern Asia (HUJV, MLAV, RESTV, XILV), South-eastern Asia (RESTV), Eastern and Southern Europe (LLOV), and Western Europe (FIWIV, KNDV, OBLV). Naturally infected hosts of filovirids are bats (BOMV and likely also other orthoebolaviruses, LLOV, MARV, MLAV, RAVV), actinopterygian fish (FIWIV, HUJV, KNDV, OBLV, XILV), domestic pigs (RESTV), nonhuman primates (MARV, TAFV), humans (BDBV, EBOV, MARV, RAVV, SUDV, TAFV), and viperid snakes (TAPV) (Negredo et al., 2011, Amman et al., 2017, Goldstein et al., 2018b, Kemenesi et al., 2018, Shi et al., 2018, Yang et al., 2019, Kuhn et al., 2020, Hierweger et al., 2021, Horie 2021, Koundouno et al., 2022, Makenov et al., 2023).
Antigenicity
Due to the absence of replicating dianlovirus, oblavirus, striavirus, tapjovirus, and thamnovirus isolates, pan-filovirid antigenicity studies have not been performed.
Derivation of names
antennarii: from genus Antennarius to which the presumed hosts of Xīlǎng virus, striated frogfish, have been assigned (Shi et al., 2018).
bombaliense: from Bombali District, Sierra Leone, where Bombali virus was discovered (Goldstein et al., 2018a).
bothropis: from genus Bothrops to which the presumed hosts of Tapajós virus, fer-de-lances, have been assigned (Horie 2021).
bundibugyoense: from Bundibugyo District, Uganda, where Bundibugyo virus was discovered (Towner et al., 2008).
Cuevavirus: from Cueva del Lloviu, a cave in Asturias Principality, Spain, where Lloviu virus was first discovered (Negredo et al., 2011).
Dianlovirus: from 滇 [diān], an abbreviation denoting China’s Yúnnán Province, and filovirid (Yang et al., 2017).
Filoviridae: from the Latin filum, “thread,” referring to the morphology of filovirid particles.
kanderense: from Kander River in Bernese Oberland, Switzerland, where Kander virus was discovered (Kiley et al., 1982).
lloviuense: from Cueva del Lloviu, a cave in Asturias Principality, Spain, where Lloviu virus was first discovered (Negredo et al., 2011).
marburgense: from Marburg an der Lahn, the city in West Germany where the first registered outbreak of Marburg virus disease occurred (Siegert et al., 1967).
menglaense: from 勐腊县 (Měnglà County), Yúnnán Province, China, where Měnglà virus was discovered (Yang et al., 2019).
Oblavirus: from Bernese Oberland, Switzerland, where Oberland virus was first discovered (Hierweger et al., 2021).
Orthoebolavirus: from the Greek ὀρθός (orthós), meaning “straight, right, proper” and the Ebola (Legbala) River in Zaire/Democratic Republic of the Congo, where the first registered outbreak of Ebola virus disease occurred (Henry 2015).
Orthomarburgvirus: from the Greek ὀρθός (orthós), meaning “straight, right, proper” and Marburg an der Lahn, the city in West Germany where the first registered outbreak of Marburg virus disease occurred (Siegert et al., 1967).
percae: from genus Perca to which the presumed hosts of Fiwi and Oberland viruses, European perch, have been assigned (Hierweger et al., 2021).
restonense: from Reston, Virgina, USA, where Reston virus was discovered (Jahrling et al., 1990).
Striavirus: from Antennarius striatus, the fish species to which the presumed hosts of Xīlǎng virus, striated frogfish, have been assigned (Shi et al., 2018).
sudanense: from Sudan, where Sudan virus was discovered (Bowen et al., 1977).
taiense: from Taï Forest, Côte d’Ivoire, where Taï Forest virus was discovered (Le Guenno et al., 1995).
Tapjovirus: from Tapajós National Forest, Brazil, where Tapajós virus was discovered (Horie 2021).
thamnaconi: from genus Thamnaconus to which the presumed host of Huángjiāo virus, horse-face filefish, has been assigned (Shi et al., 2018).
Thamnovirus: from Thamnaconus septentrionalis, the fish species to which the presumed host of Huángjiāo virus, greenfin horse-faced filefish, has been assigned (Shi et al., 2018).
zairense: from Zaire (today Democratic Republic of the Congo), where Ebola virus was discovered (Johnson et al., 1977).
Genus demarcation criteria
PAirwise Sequence Comparison (PASC) using coding-complete filovirid genomes is the primary tool for filovirid genus demarcation. Genomic sequences of filovirids of different genera differ from each other by ≥55% (Bào et al., 2017). Genomic features, such as the number and location of gene overlaps, the number of open reading frames (ORFs) and/or genes, filovirid host and geographic distribution, and filovirid pathogenicity for different organisms are also taken into account for genus assignment.
Relationships within the family
Phylogenetic relationships across the family have been established from maximum likelihood trees generated using coding-complete or complete genome sequences (Figure 4.Filoviridae) or by phylogenetic analysis of RdRP sequences (Wolf et al., 2018).
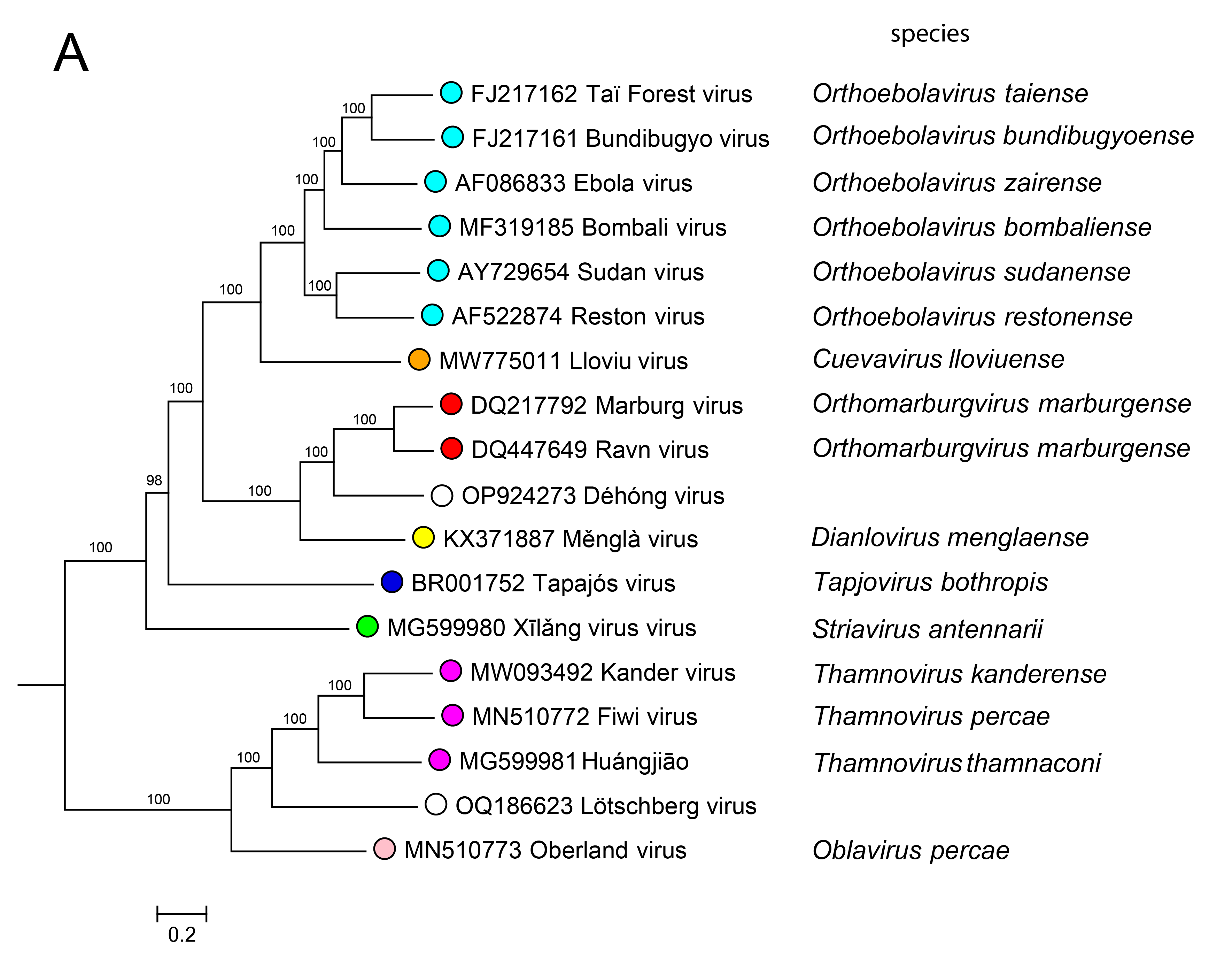 |
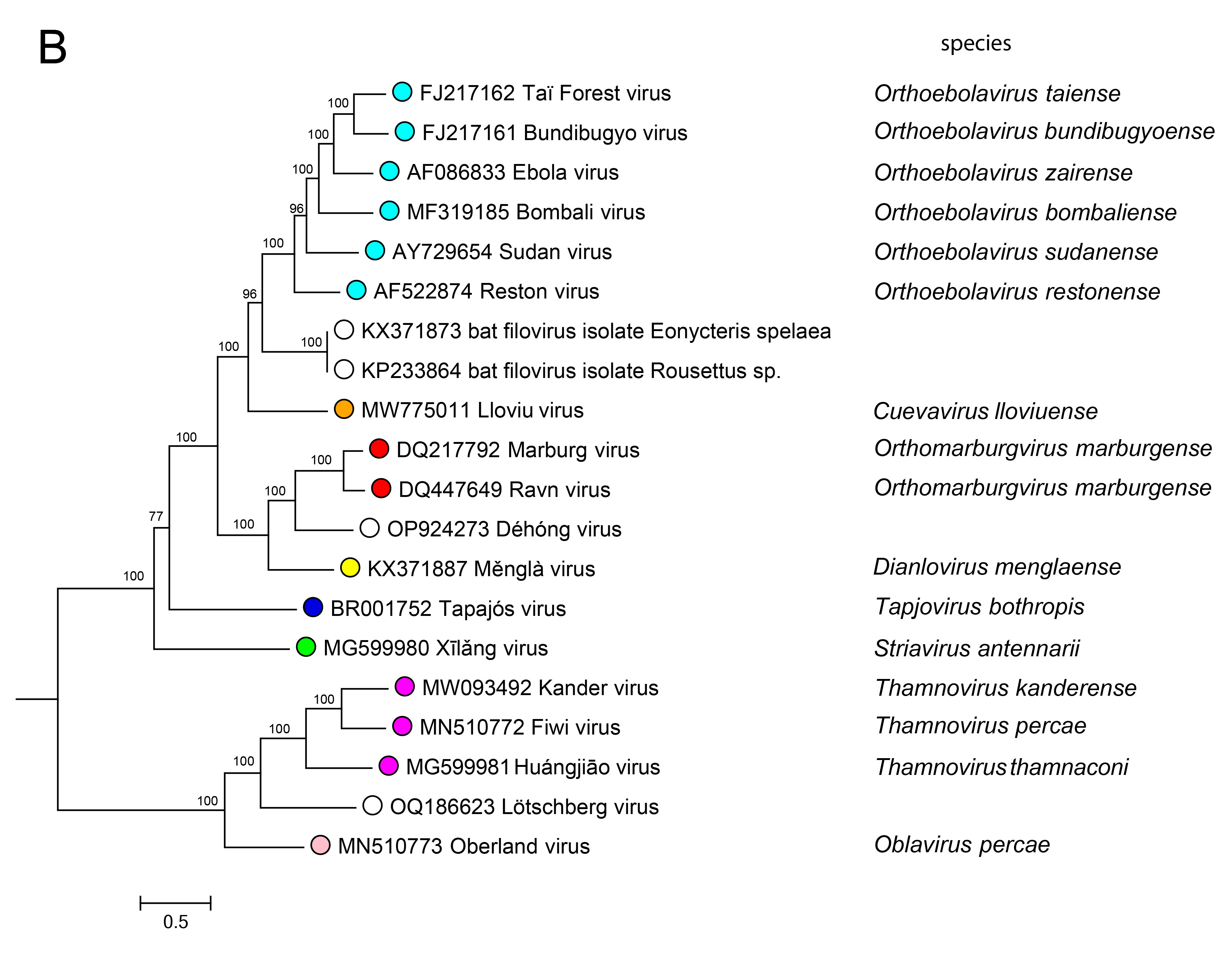 |
| Figure 4. Filoviridae. Phylogenetic relationships of filovirids. Maximum-likelihood tree (midpoint-rooted) inferred by using A) coding-complete or complete filovirus genome sequences and (B) filovirus RNA-directed RNA polymerase gene (L) sequences. Sequences were translationally aligned using Clustal-Omega version 1.2.3 (http://www.clustal.org/omega/) and were manually curated in Geneious version R9 (http://www.geneious.com) or Unipro UGENE version 35 (http://ugene.net/). Trees were inferred in FastTree version 2.1 (http://www.microbesonline.org/fasttree/) using a General Time Reversible (GTR) model with 20 Gamma-rate categories, 5,000 bootstrap replicates, and exhaustive search parameters (-slow) and pseudocounts (-pseudo). Numbers near nodes on the trees indicate bootstrap values in percentages. Tree branches are scaled to nucleotide substitutions per site (scale bars). |
Relationships with other taxa
Filovirids are closely related to other viruses in the order Mononegavirales, in particular paramyxovirids, pneumovirids, and sunvirids (Wolf et al., 2018).
Related, unclassified viruses
Virus names and abbreviations are not official ICTV designations.
*Coding region sequence incomplete.
Additional unclassified filovirids that are probable members of existing genera are listed under individual genus descriptions.

