Family: Cruliviridae
Jens H. Kuhn, Scott Adkins, Katherine Brown, Juan Carlos de la Torre, Michele Digiaro, Holly R. Hughes, Sandra Junglen, Amy J. Lambert, Piet Maes, Marco Marklewitz, Gustavo Palacios, Takahide Sasaya (笹谷孝英), Massimo Turina, and Yong-Zhen Zhang (张永振)
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Yong-Zhen Zhang (张永振) (zhangyongzhen@fudan.edu.cn)
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023, updated June 2024
Summary
Cruliviridae is a family of negative-sense RNA viruses with genomes of 10.8–11.5 kb (Table 1 Cruliviridae). These viruses have been found in crustaceans. The family includes a single genus with three species for three viruses. The genomes of crulivirids consist of three RNA segments with open reading frames (ORFs) that encode nucleoproteins (NPs), glycoproteins (GPs), large (L) proteins containing RNA-directed RNA polymerase (RdRP) domains, and, in some members of the family, zinc-finger (Z) proteins of unknown function.
Table 1 Cruliviridae. Characteristics of members of the family Cruliviridae
| Characteristic | Description |
| Example | Wēnlǐng crustacean virus 9 (S: KX884858; M: KX884857; L: KX884856), species Lincruvirus wenlingense, genus Lincruvirus |
| Virion | Enveloped, spherical |
| Genome | 10.8–11.5 kb of tri-segmented negative-sense RNA |
| Replication | Largely undefined |
| Translation | Unknown |
| Host range | Portunid crustaceans (crabs) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Bunyaviricetes, order Elliovirales; the family includes one genus and four species |
Virion
Morphology
Crulivirids produce enveloped spherical particles (Bojko et al., 2018, Bojko et al., 2019).
Nucleic acid
Crulivirid genomes have three RNA segments (small [S], medium [M], and large [L]) of linear negative-sense RNA with a total length of 10.8–11.5 kb (S segment: about 0.8–1.6 kb; M segment: about 3.2–3.6 kb; and L segment: about 6.7 kb). (Shi et al., 2016, Bojko et al., 2019, Huang et al., 2019).
Genome organization and replication
Viruses of the family Cruliviridae each have at least one ORF that encodes an NP, a GP, an L protein containing an RdRP domain (Figure 1 Cruliviridae). Some viruses also encode a further zinc-finger (Z) protein of unknown function (Shi et al., 2016, Bojko et al., 2019, Huang et al., 2019). Individual genomic segments are flanked by conserved sequences (Bojko et al., 2019). Replication begins with endocytosis of enveloped virions. Infected host cells develop small and large vesicles containing progeny particles in various stages of development. Expansion of large vesicles precede the release of smaller vesicles containing mature virions into the cytoplasm before exiting the cell (Bojko et al., 2018, Bojko et al., 2019).
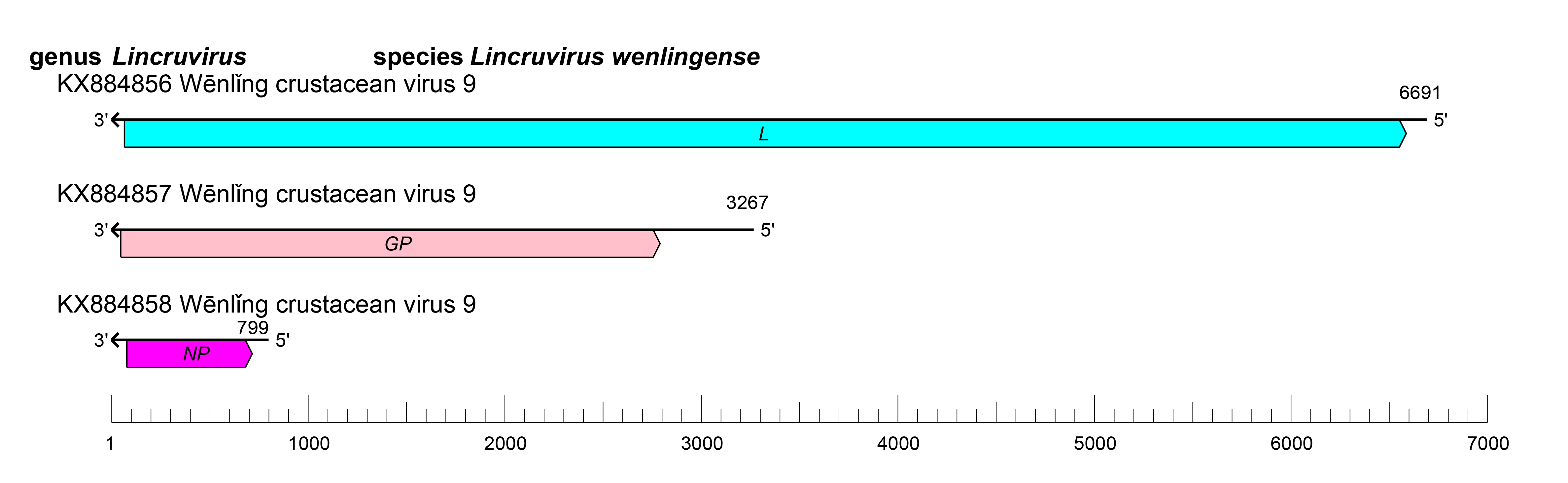 |
| Figure 1.Cruliviridae. Genome organization of a representative virus of family Cruliviridae, Wēnlǐng crustacean virus 9. ORFs are colored according to the predicted protein function (GP, glycoprotein gene; L, large protein gene; NP, nucleoprotein gene). |
Biology
The four classified crulivirids have been detected in crustaceans:
- Chinese mitten crab virus 1 (CmcV1; previously called Eriocheir sinensis bunya like virus) was discovered in Chinese mitten crabs (portunid Eriocheir sinensis Milne-Edwards, 1853) (Huang et al., 2019) in China
- European shore crab virus 1 (EscV1; previously called Carcinus maenas portunibunyavirus 1 [CmPBV1]) was discovered in European shore crabs (portunid Carcinus maenas (Linnaeus, 1758)) of Faroe Islands, Denmark (Bojko et al., 2019)
- Wēnlǐng crustacean virus 9 (WlCV9) was discovered in unspecified crustaceans in China (Shi et al., 2016)
- Callinectes danae portunibunyavirus 1 (CdPBV-1) was discovered in the swimming crab Callinectes danae S. I. Smith, 1869 in Brazil
Unclassified crulivirids have been detected in European brown shrimp (crangonid Crangon crangon (Linnaeus, 1758)) in Belgium (Van Eynde et al., 2020).
Derivation of names
braziliense: from Brazil
Cruliviridae: from Crustacean lincruvirus
europense: from Europe;
Lincruvirus: from Wēnlǐng crustacean virus 9
sinense: from Latin sinae, meaning “China”
wenlingense: from Wēnlǐng (温岭市), China
Genus demarcation criteria
Not applicable (the family includes only a single genus).
Species demarcation criteria
Members of different species are phylogenetically distinct upon analysis of the L protein nucleotide sequence.
Relationships within the family
Phylogenetic relationships of members of the family Cruliviridae are shown in Figure 2 Cruliviridae.
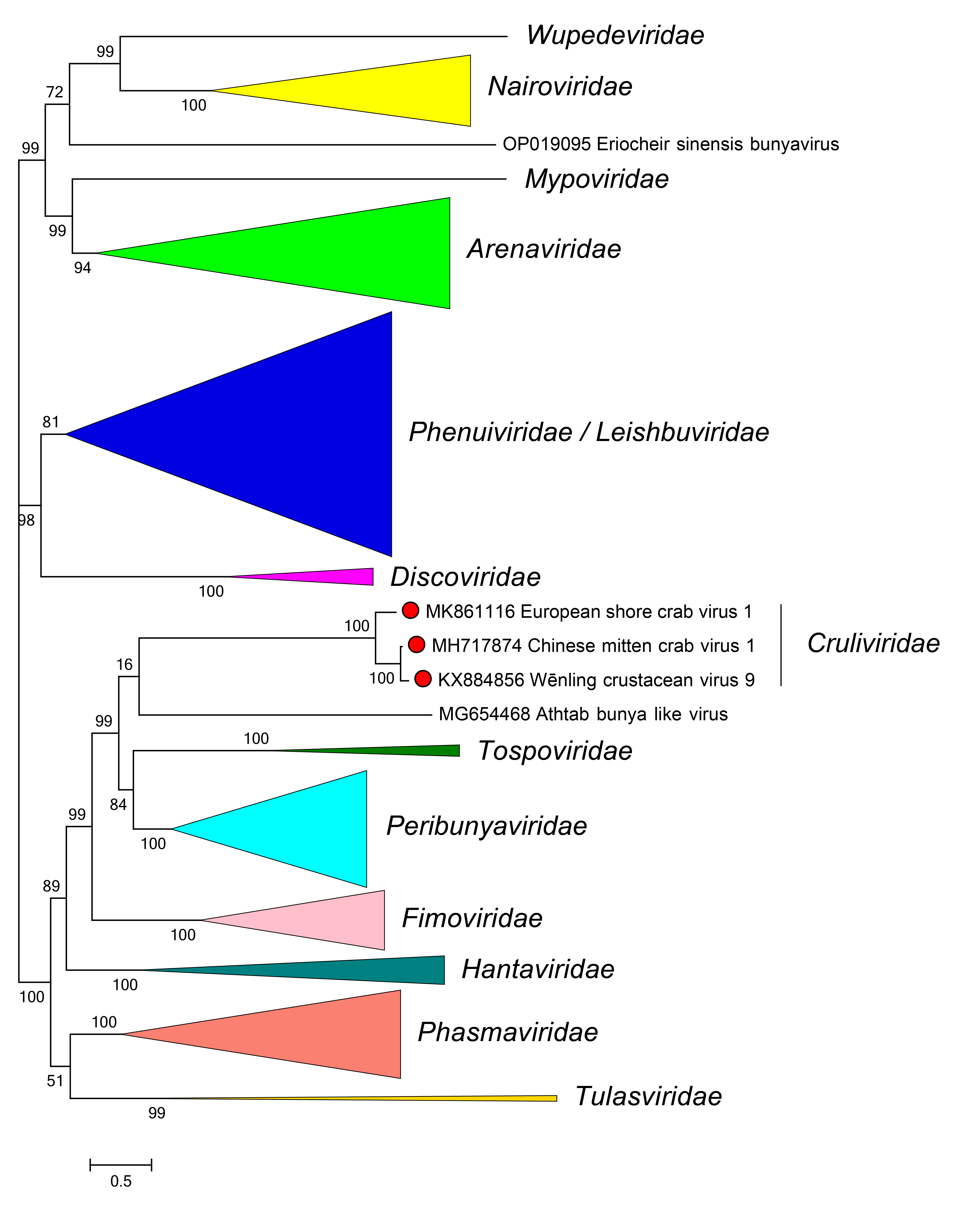 |
| Figure 2 Cruliviridae. Phylogenetic relationships of viruses in the family Cruliviridae. |
Branches for viruses in other families are collapsed. L protein sequences were aligned using MUSCLE and a maximum likelihood tree was produced using FastTree with default settings. Numbers at nodes indicate bootstrap support where this was >70%. Details of the virus sequences used are available in the treefile and alignment file in the Resources section.
Relationships with other taxa
Viruses in the family Cruliviridae are most closely related to bunyaviral fimovirids, peribunyavirids, phasmavirids, tulasvirids, and tospovirids (Herath et al., 2020, Olendraite et al., 2023).

