Family: Chuviridae
Jens H. Kuhn, Nolwenn M. Dheilly, Sandra Junglen, Sofia Paraskevopoulou (Σοφία Παρασκευοπούλου), Mang Shi (施莽) and Nicholas Di Paola
The citation for this ICTV Report chapter is the summary published as:
Corresponding author: Nicholas Di Paola (nicholas.dipaola.civ@health.mil)
Edited by: Jens H. Kuhn and Stuart G. Siddell
Posted: November 2023
Summary
Chuviridae is a family of negative-sense RNA viruses with genomes of 9.1–12.2 kb (Table 1.Chuviridae). These viruses have been associated with arachnids, barnacles, crustaceans, insects, fish, and reptiles in Africa, Asia, Australia, Europe, North America, and South America. The family includes 16 genera and 43 species for 44 viruses. The chuvirid genome contains two to four open reading frames (ORFs) that encode a glycoprotein (GP), a nucleoprotein (NP), a large (L) protein containing an RNA-directed RNA polymerase (RdRP) domain, and/or proteins of unknown function.
Table 1.Chuviridae Characteristics of members of the family Chuviridae
| Characteristic | Description |
| Example | Wēnzhōu crab virus 2 (segment 1: KM817601; segment 2: KM817602), species Chuvivirus canceris, genus Chuvivirus |
| Virion | Unknown |
| Genome | 9.1–12.2 kb of nonsegmented linear, nonsegmented circular, bisegmented linear or bisegmented circular negative-sense RNA |
| Replication | Unknown |
| Translation | Unknown |
| Host range | Araneaen and ixodidan arachnids (spiders and ticks); barnacles; decapod crustaceans (crabs and shrimp); blattodean (cockroach), coleopteran (glow-worm), dermapteran (earwig), dipteran (louse fly, mosquito), hemipteran (stink bug, water bug, whitefly), hymenopteran (wasp), odonatan (damselfly/dragonfly), and polyneopteran (termite) insects; atheriniform and syngnathiform fish; and squamate reptiles (snakes) |
| Taxonomy | Realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, class Monjiviricetes, order Jingchuvirales; the family includes 16 genera and 43 species for 44 viruses |
Virion
Morphology
Unknown.
Nucleic acid
Chuvirids have nonsegmented linear, nonsegmented circular, bisegmented linear or bisegmented circular negative-sense RNA genomes with total lengths of 9.1–12.2 kb (Li et al., 2015, Hang et al., 2016, Harvey et al., 2019, Käfer et al., 2019, Argenta et al., 2020, Gondard et al., 2020, Chang et al., 2021, Costa et al., 2023) (Shi et al., 2016, Sadeghi et al., 2018, Tokarz et al., 2018, Sameroff et al., 2019, Viljakainen et al., 2020, Wu et al., 2020).
Genome organization and replication
Chuvirid genomes have two to four ORFs that encode a glycoprotein (GP), a nucleoprotein (NP), a large (L) protein, and/or proteins of unknown function (Li et al., 2015, Hang et al., 2016, Harvey et al., 2019, Käfer et al., 2019, Argenta et al., 2020, Gondard et al., 2020, Chang et al., 2021, Costa et al., 2023) (Shi et al., 2016, Sadeghi et al., 2018, Tokarz et al., 2018, Sameroff et al., 2019, Viljakainen et al., 2020, Wu et al., 2020). The replication cycle of chuvirids remains to be elucidated.
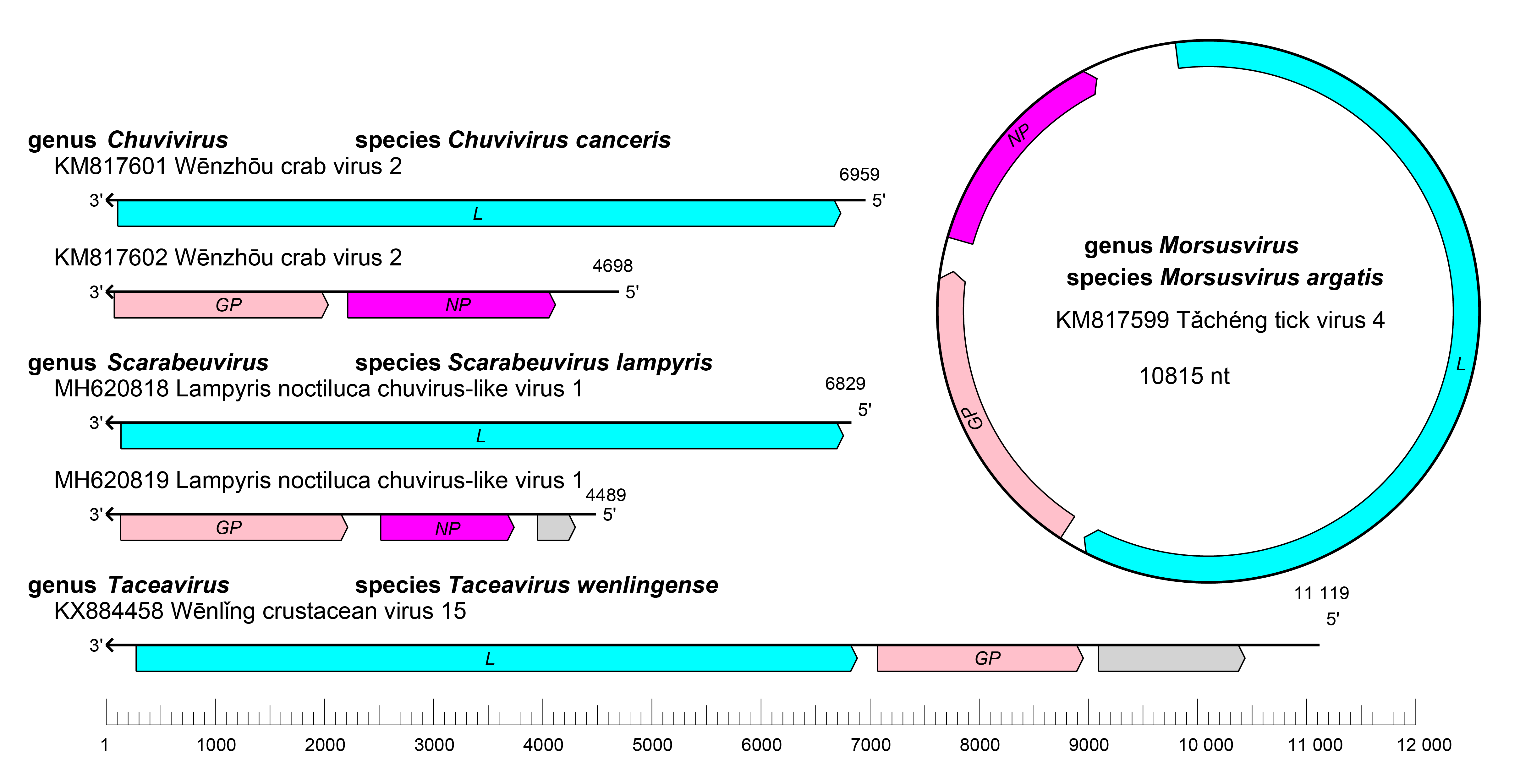 |
| Figure 1.Chuviridae. Genome organization of four representative viruses in the family Chuviridae. ORFs are colored according to the predicted protein function. GP, glycoprotein gene; NP; nucleoprotein gene; L, large protein gene. Grey color indicates ORFs encoding proteins of unknown function. |
Biology
Classified chuvirids have been associated with araneaen and ixodidan arachnids (spiders and ticks); barnacles; decapod crustaceans (crabs and shrimp); blattodean (cockroach), coleopteran (glow-worm), dermapteran (earwig), dipteran (louse fly, mosquito), hemipteran (stink bug, water bug, whitefly), hymenopteran (wasp), and odonatan (damselfly/dragonfly) insects; atheriniform and syngnathiform fish; and squamate reptiles (snakes).
In addition, unclassified chuvirids have been associated with cephalopods (Rey-Campos et al., 2023), termites (Litov et al., 2022) and turtles (Laovechprasit et al., 2023); at least one unpublished report suggests that chuvirids may infect humans (Quan et al., 2020). Endogenized chuvirid elements (in particular G-gene-derived nucleic acids) in arthropod genomes indicate that chuvirids are millions of years old (Dezordi et al., 2020, Dezordi et al., 2023).
Derivation of names
amblyommae: from the host genus Amblyomma;
argatis: from the host genus Argas;
belostomatis: from Belostoma flumineum mononega-like virus;
blattae: from the host genus Blatta;
boleense: from Bólè tick virus 3;
Boscovirus: from Medieval Latin boscus, meaning “wood
brunnichi: from Brünnich
canceris: from the Latin cancer, meaning “crab”
chalcocoris: from Chalcocoris rutilans mononega-like virus 2
changpingense: from Chāngpíng tick virus 2
chulinense: from hymenopteran chu-related virus OKIAV147
Chuviridae: from the ancient Chinese 楚 (Chǔ) State during the Spring and Autumn
Chuvivirus: from Jingchuvirales
cirripedis: from the host genus Cirripedia
craterocephali: from the host genus Craterocephalus
culicidae: from the host genus Culex
Culicidavirus: from culicid, derived from host genus Culex
culicis: from culicid, derived from host genus Culex
culisetae: from the host genus Culiseta
dentati: from the Latin dentātī, meaning “tooth”
dermacentoris: from the host genus Dermacentor
Demapteravirus: incompletely from the Greek derma, meaning “skin”; pteron, meaning “‘wing”;
dermapteri: from dermapteran chu-related virus 142
Doliuvirus: from the Latin dōlium, meaning “cask”
draconis: from the Ancient Greek δράκων (drákōn), meaning “serpent, dragon”
fabricii: after Fabricius
franki: from Herr Frank virus 1
genovaense: from Genoa virus
hippoboscidae: from thehost family Hippoboscidae
hubeiense: from Húběi chuvirus-like virus 1
hypoboscidae: from the host family Hippoboscidae
imjinense: from Imjin River virus 1
ixodes: from the host genus Ixodes
karukeraense: from Karukera tick virus
lampyris: from Lampyris noctiluca chuvirus-like virus 1
lishiense: from Lĭshì spider virus 1
lycodontis: from the host genus Lycodon
Mivirus: from the Chinese 芈 (Mǐ), the ancestral name of King Zhuang of Chǔ State during the Spring and Autumn Period
Morsusvirus: from the Proto-Italic *mordeō, meaning “I bite”,
Nigecruvirus: from the Latin niger meaning “black” and the Latin cru meaning “leg”, referrering to blacklegged tick chuvirus-2 which was discovered by HTS of the deer tick Ixodes scapularis Say, 1821
Odonatavirus: from the host order Odonata
odontis: from odonatan chu-related virus 136
Pediavirus: from the Ancient Greek πεδίον (pedíon), meaning “field”
periplanetae: from Periplaneta americana mononega-like virus
Piscichuvirus: from the Latin piscis, meaning “fish”; chuvirus
Pterovirus: from the Greek pteron, meaning “wing”
quitotaense: from Quitota
rhipicephali: from the host genus Rhipicephalus
Rochuvirus: from Proto-Indo-European *pro-, meaning “forth, forward”; chuvirus
sanxiaense: from Sānxiá atyid shrimp virus 4
Scarabeuvirus: from scarab beetle and -virus: suffix for a virus genus
suffolkense: from Suffolk virus
supellae: from Supella longipalpa mononega-like virus 2
Taceavirus: from the Latin tacēre, meaning “to keep quiet”
trialeurodis: from Trialeurodes vaporariorum mononega-like virus 2
trichopriae: from Trichopria drosophilae mononega-like virus
Vapochuvirus: from the Latin vapor, meaning “steam, heat”
wenlingense: Wēnlǐng fish chu-like virus and Wēnlǐng crustacean virus 15
wuhanense: from Wǔhàn tick virus
Genus demarcation criteria
Viruses in different chuvirid genera have L protein sequences with <31% amino-acid identity (Di Paola et al., 2022).
Species demarcation criteria
Viruses in different chuvirid species have L protein sequences with <90% amino-acid identity (Di Paola et al., 2022).
Relationships within the family
Phylogenetic relationships of members of the family Chuviridae are shown in Figure 2.Chuviridae. In this analysis, viruses from the genus Mivirus are not monophyletic; this anomaly will be addressed in a future taxonomic proposal.
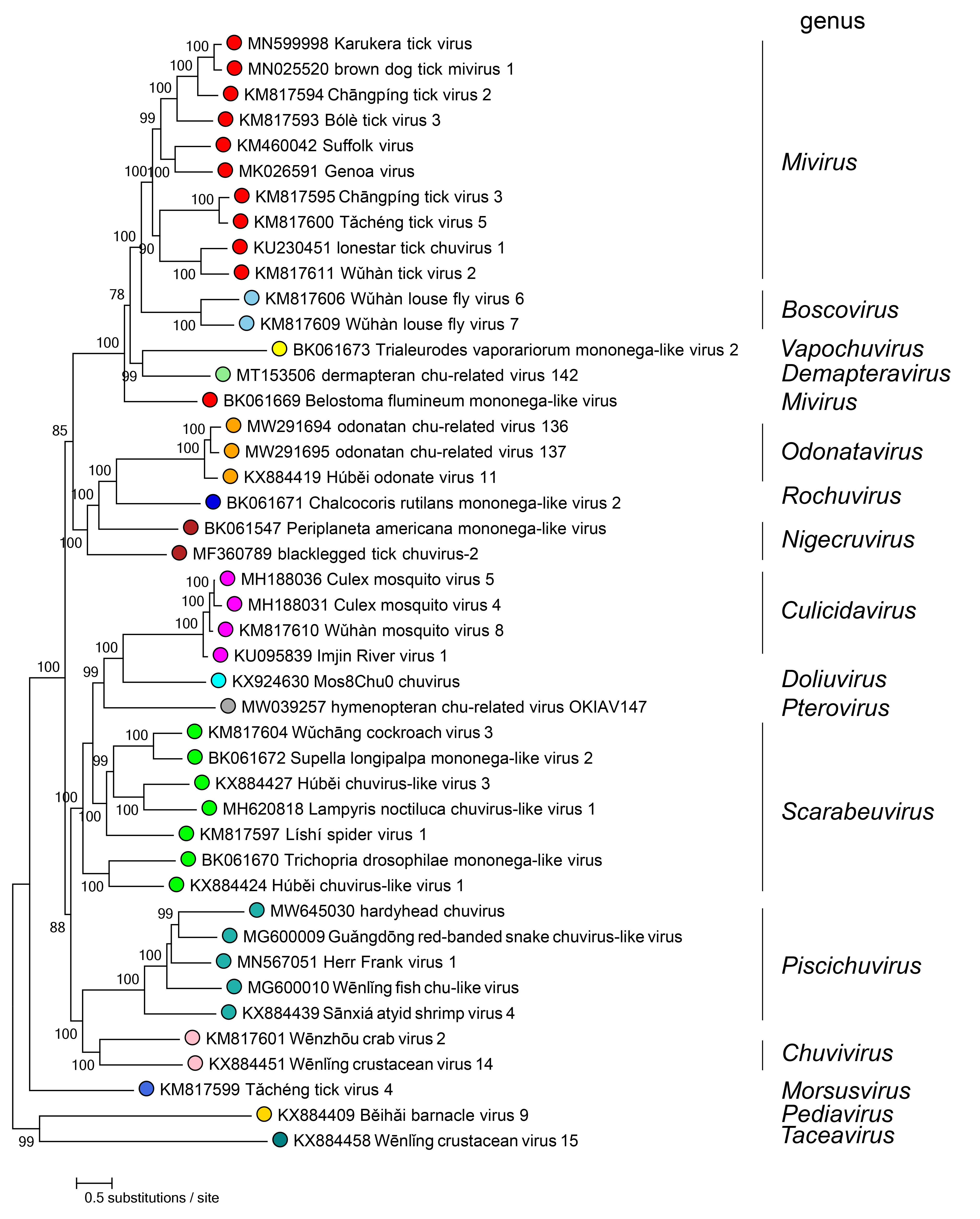 |
| Figure 2.Chuviridae. Phylogenetic relationships of chuvirids. Maximum-likelihood tree (midpoint-rooted) inferred by using large protein gene (L) sequences. Sequences were initially aligned by MAFFT version 7 (https://mafft.cbrc.jp/alignment/software/) in Geneious version R9 (http://www.geneious.com) and realigned using ClustalW (https://www.genome.jp/tools-bin/clustalw). The tree was estimated using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/), a subtree pruning and regrafting (SPR) topology searching algorithm, and a Bayes branch support algorithm. Numbers near nodes on the trees indicate bootstrap values in decimal form. Tree branches are scaled to nucleotide substitutions per site (scale bar). |
Relationships with other taxa
The viruses in the family Chuviridae are most closely related to jingchuviral aliusvirids, crepuscuvirids, myriavirids and natarevirids (Di Paola et al., 2022).
Related, unclassified viruses
| Virus name | Accession number | Virus abbreviation | Reference |
| Atrato chu-like virus 1 | MN661027* | ||
| Chuviridae sp. isolate CA | MZ771201* | (Konstantinidis et al., 2022) | |
| Coleopteran chu-related virus OKIAV151 | MT153523* | (Käfer et al., 2019) | |
| grapevine-associated jingchu-like virus 1 | MW648445* | Nerva et al., 2022) | |
| haranti vitus /Chuviridae sp. isolate GA | MZ771218; MZ771219* | (Konstantinidis et al., 2022) | |
| midge associated chuvirus M1C4 | LR701643* | (Modha et al., 2019) | |
| Schistocephalus solidus jingchuvirus | MN803434* | (Hahn et al., 2020) | |
Tasmanian devil chu-like virus | (Harvey et al., 2023) |
Virus names and virus abbreviations are not official ICTV designations.
* Incomplete genome

