Family: Adenoviridae
Mária Benkő, Koki Aoki, Niklas Arnberg, Andrew J. Davison, Marcela Echavarría, Michael Hess, Morris S. Jones, Győző L. Kaján, Adriana E. Kajon, Suresh K. Mittal, Iva I. Podgorski, Carmen San Martín, Göran Wadell, Hidemi Watanabe and Balázs Harrach*
The citation for this ICTV Report chapter is the summary published as Benkő et al., (2022):
ICTV Virus Taxonomy Profile: Adenoviridae 2022, Journal of General Virology 103(3):001721
Corresponding author: Balázs Harrach (balazs.harrach@gmail.com)
Edited by: Arvind Varsani and Andrew J. Davison
Posted: November 2021, updated August 2024
Summary
The family of Adenoviridae consists of viruses with non-enveloped, icosahedral virions containing linear dsDNA genomes of 25–48 kb (Table 1 Adenoviridae). Its members infect a variety of vertebrate hosts ranging from fish to humans and are allocated to six genera. Members of genus Mastadenovirus infect mammals, those of genus Aviadenovirus infect birds, genus Ichtadenovirus contains a single fish adenovirus, and strains of genus Testadenovirus occur in turtles. Members of genus Barthadenovirus (formerly Atadenovirus) occur in squamate reptiles, birds, ruminants, marsupials and tortoises, and those of genus Siadenovirus infect birds, frog and tortoise. The severity of infections ranges from subclinical to lethal. Adenoviruses are popular virus vectors, for example in vaccination against the coronavirus SARS-CoV-2.
Table 1 Adenoviridae. Characteristics of members of the family Adenoviridae
| Characteristic | Description |
| Example | human adenovirus 5 (AC_000008), species Mastadenovirus caesari, genus Mastadenovirus |
| Virion | Non-enveloped icosahedral capsids, 90 nm in diameter |
| Genome | Linear, dsDNA of 25–48 kb with inverted terminal repeats |
| Replication | Nuclear |
| Translation | From capped, polyadenylated and often spliced transcripts |
| Host range | Mammals, birds, reptiles, amphibians and fish; host range varies among genera |
| Taxonomy | Realm Varidnaviria, kingdom Bamfordvirae, phylum Preplasmiviricota, subphylum Polisuviricotina, class Pharingeaviricetes, order Rowavirales; 6 genera containing 125 species |
Virion
Morphology
Adenovirus (AdV) virions are non-enveloped particles, 90 nm in diameter. The characteristic icosahedral capsid has a pseudo T=25 triangulation number and consists of 240 non-vertex capsomers (hexons), 8–10 nm in diameter, and 12 vertex capsomers (penton bases), from which fibers protrude (Figure 1 Adenoviridae) (van Raaij et al., 1999). The 240 hexons are formed by the interaction of three identical proteins (designated II) and consist of two distinct parts: a triangular top with three “towers”, and a pseudohexagonal base with a central cavity. The hexon bases are tightly packed, forming a protein shell that protects the inner components. The 12 penton bases are each formed by the interaction of five copies of protein III and are tightly associated with the fibers, which are homotrimers of protein IV. The fibers have three structural domains, including the tail, which binds to the penton base, the shaft of characteristic length and the distal knob (San Martín 2012). The length of fibers examined ranges from 9 to 77.5 nm. Human adenovirus 40 (HAdV-40) and human adenovirus 41 (HAdV-41) have fibers of two different lengths that occur alternately on the vertices. Human adenovirus 52 and many simian adenoviruses (SAdVs) also have fibers of two different lengths (Jones et al., 2007, Lenman et al., 2015), and some SAdVs even have three different fibers (Abbink et al., 2015, Podgorski et al., 2023). Members of genera Aviadenovirus and Barthadenovirus may have more than one fiber inserted at a single vertex (Gelderblom and Maichle-Lauppe 1982, Pénzes et al., 2014). In members of genus Mastadenovirus, protein IX is located on the outer part of the capsid, cementing the hexon proteins in each facet (12 copies of IX per facet). In some human adenoviruses (HAdVs), protein IX also links two facets together across the icosahedral edge (Liu et al., 2010). In HAdV-41, the organization of the external minor coat protein IX is different from that of all previously characterized human and nonhuman mastadenoviruses, and some partially ordered elements were identified reinforcing the vertex region. This suggests a role in enhancing the physicochemical capsid stability of this enteric adenovirus to withstand the harsh conditions found in the gut (Pérez-Illana et al., 2021, Rafie et al., 2021). Protein IX is not present in the members of the other genera. A barthadenovirus-specific protein, LH3, has a similar structural role to IX, though a distantly related structure (Gorman et al., 2005, Pantelic et al., 2008). Proteins IIIa, VI and VIII are located internally. Five IIIa monomers are arranged in a ring underneath each vertex (penton base). Protein VI is located beneath the hexons and is not arranged in icosahedral symmetry. There are six monomers of protein VIII per facet: three are wedged between protein IIIa and the peripentonal hexons, and the other three are arranged around the icosahedral threefold symmetry axis, contributing with protein IX to the stabilization of the facet (Liu et al., 2010). The core consists of the DNA genome complexed with four proteins: V, VII, X (also known as mu or µ) and terminal protein (TP). Protein V is found only in mastadenoviruses. A few copies of the maturation protease (L3 23K, adenovirus protease/AVP/adenain) are packaged bound to the genome (Mangel and San Martín 2014).
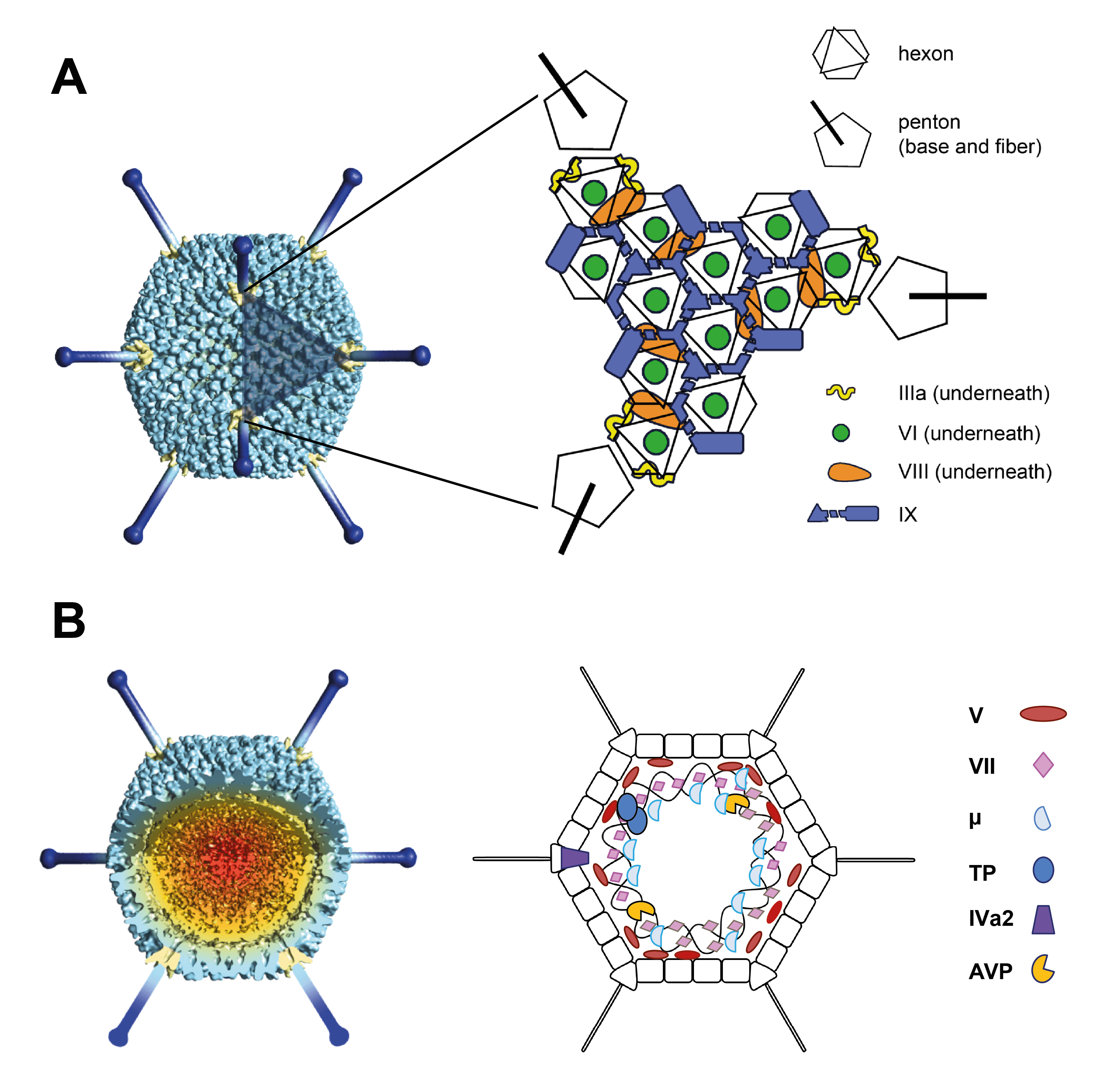 |
| Figure 1 Adenoviridae (A) The icosahedral adenovirus capsid. Left: Model prepared by low resolution cryo-electron microscopy with penton bases highlighted in yellow and fibers in dark blue (from (van Raaij et al., 1999). The shaded triangle indicates one facet. Right: Schematic figure indicating the position of the major and minor coat proteins in the facet, as solved by cryo-electron microscopy of the mastadenovirus human adenovirus 5 (Liu et al., 2010). (B) Non-icosahedral components. A segment has been removed from the map in (A) to reveal the core. For detailed description of the capsid and core proteins see text. As the structure of the nucleoprotein core has not been established, the polypeptides associated with the DNA are shown in hypothetical locations. Panels A and B reproduced from (San Martín 2012). |
Physicochemical and physical properties
Virion Mr is 150–180×106; and buoyant density in CsCl is 1.31–1.36 g cm−3. Viruses are stable on storage in the frozen state. They are insensitive to lipid solvents. Human adenovirus 5 (HAdV-5) partially disassembles at pH 6. Heat sensitivity varies among genera (Bartha and Kisary 1970).
Nucleic acid
The genome is a single, linear molecule of dsDNA that contains inverted terminal repeats (ITRs) at the ends. TP is covalently linked to the 5′-end of each DNA strand. The size of the genomes sequenced to date ranges between 24,630 and 48,395 bp, with ITRs of 26–721 bp. The G+C content of the genome varies widely between 31.3 and 70.0%. The genetic organization of the central part of the genome is well conserved throughout the family, whereas the regions near the ends exhibit large variations in length and gene content (Figure 2 Adenoviridae) (Davison et al., 2003b, Doszpoly et al., 2019).
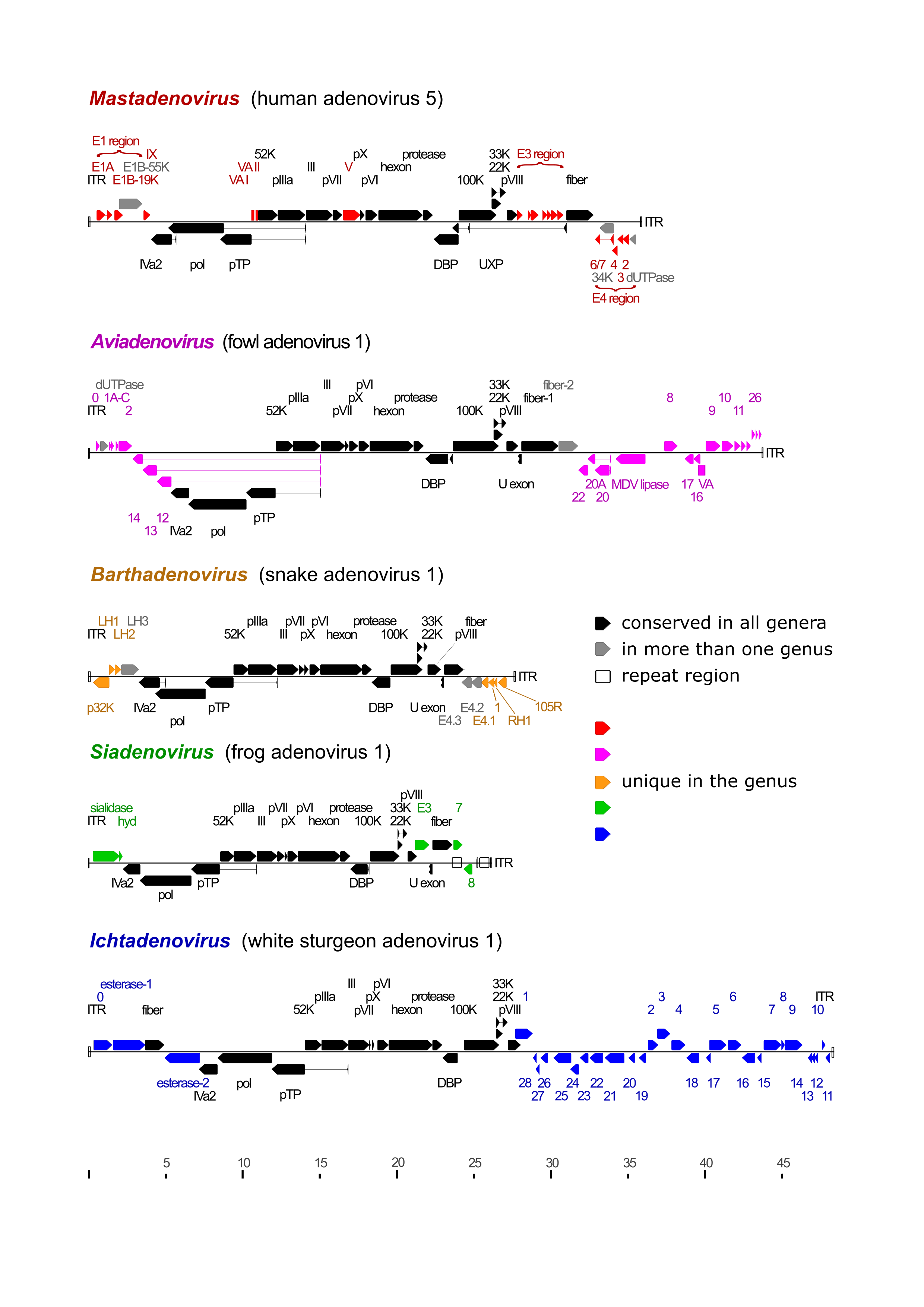 |
| Figure 2 Adenoviridae Schematic illustration of the various genome organizations found in representative members of the adenovirus genera. Black arrows depict coding sequences conserved in every genus, grey arrows show those present in more than one genus, and coloured arrows show coding sequences of genus-specific proteins. Proteins without identified function are named according to ORF number, but the ORF prefix is omitted for clarity. White rectangles show direct repeat regions and inverted terminal repeats (ITRs; not clearly visibly when very short). Thin lines connect the exons of spliced genes. Aviadenovirus (fowl adenovirus 1) ORF2, ORF12, ORF13 and ORF14 are homologues of parvovirus (family Parvoviridae) NS1, MDV lipase is homologous to Marek's disease virus (family Orthoherpesviridae) lipase, and white sturgeon adenovirus 1 has two different homologs of Escherichia coli O-acetylesterase. |
Proteins
About 40 different viral polypeptides are produced (Table 2 Adenoviridae). Almost one-third of these compose the virion, including a virus-encoded cysteine protease (23 kDa) that is necessary for the processing of some precursor proteins (prefixed with p) (Mangel and San Martín 2014). With the exception of proteins V and IX, the structural proteins are well conserved in every genus (Davison et al., 2003b, Doszpoly et al., 2019).
Table 2 Adenoviridae. Proteins encoded by human adenovirus 2
| kDa | Transcription class | Description | Note |
|---|---|---|---|
| 13, 27, 32 | E1A | R | Only in mastadenoviruses |
| 16, 21 | E1B | R | Only in mastadenoviruses |
| 55 | E1B | R | Only in mastadenoviruses |
| 59 | E2A | D; 72 kDa* DBP | |
| 120 | E2B | D; 140 kDa* pol | |
| 75 | E2B | D, S; 87 kDa* pTP† | |
| 4, 7, 8, 10, 12 | E3 | H | Only in mastadenoviruses |
| 13, 15, 15, 19 | |||
| 7, 13, 13, 14 | E4 | R | Only in mastadenoviruses |
| 15 | E4 | R; 31 kDa* dUTPase | Only in some mast- and aviadenoviruses |
| 17 | E4 | R; 34 kDa* | Only in mast- and barthadenoviruses; present on the capsid at a single location |
| 47 | L1 | D;† 52/55 kDa* | |
| 64 | L1 | D, S (pIIIa);† p-protein | |
| 63 | L2 | S (III); penton base* | |
| 22 | L2 | S (pVII);† major core | |
| 42 | L2 | S (V); minor core | Only in mastadenoviruses |
| 10 | L2 | S (pX);† X/µ | |
| 27 | L3 | S (pVI)† | |
| 109 | L3 | S (II); hexon | |
| 23 | L3 | D, S; protease | |
| 90 | L4 | D; 100 kDa* | |
| 25 | L4 | D, R; 33 kDa* p-protein | |
| 22 | L4 | D, R; 22 kDa* p-protein, shares first 105 amino acid residues with L4 33K | |
| 25 | L4 | S (pVIII)† | |
| 62 | L5 | S (IV); fiber | |
| 14 | Intermediate | S (IX) | Only in mastadenoviruses |
| 51 | Intermediate | D, S (IVa2) |
Molecular masses are rounded to the nearest 1000, and are presented as unmodified and uncleaved gene products. D = DNA synthesis and packaging; DBP = DNA-binding protein; H = subverting host defence mechanisms; p = precursor; p-protein = phosphoprotein; pol = DNA polymerase; R = regulation, S = structural; TP = terminal protein; * = Mr values are significantly different from those obtained by SDS-PAGE; † = cleaved by viral protease. L4 33K is present in the empty capsid, and DBP is present both in empty and mature virions (Ahi et al., 2015).
Lipids
None reported.
Carbohydrates
Fiber proteins and some of the non-structural proteins are glycosylated.
Genome organization and replication
Virus entry into cells occurs by attachment via the fiber knob to various receptors (Arnberg 2012, Lenman et al., 2015, Lenman et al., 2018, Stasiak and Stehle 2020) and subsequent internalization via interaction between the penton base and cellular αv integrins (Smith et al., 2010, Greber and Flatt 2019, Pied and Wodrich 2019). Recent results show that alpha-defensin binding expands human adenovirus tropism by providing a novel pathway for HAdV binding to cells that bypasses viral primary receptors (Zhao et al., 2024). Protein VI mediates the release of virions from endosomes, allowing dynein-mediated transport on microtubules to nuclear pores. After uncoating, the virus core is delivered to the nucleus, which is the site of virus transcription, DNA replication and virion assembly (Hoeben and Uil 2013, Condezo and San Martín 2017). Virus infection mediates the early shut-off of host DNA synthesis and, later, synthesis of host mRNAs and polypeptide. Transcription by host RNA polymerase II involves both DNA strands of the virus genome, and initiates (in human adenovirus 2 [HAdV-2]) from promoters of the five early regions (E1A, E1B, E2, E3 and E4), two genes of intermediate proteins (IX and IVa2), the major late promoter (L) as well as the U exon protein (UXP) late promoter (Figure 3 Adenoviridae). All primary transcripts are capped and polyadenylated. Complex splicing generates families of mRNAs, and the proteins are produced via these complex splicing mechanisms (Figure 3 Adenoviridae). In primate mastadenoviruses, there are one or two virus-associated (VA) RNA genes, which are transcribed by RNA polymerase III. These encode RNA products that facilitate translation of late mRNAs and block the cellular interferon response. Similar VA RNA genes have not been identified in other adenoviruses. In some fowl adenoviruses, the existence of one VA RNA gene, at a different genome position, has been described, but these VA RNAs are not homologous to mastadenovirus VA RNAs (Chiocca et al., 1996). Products of the four early (E) regions of mastadenovirus genomes (E1 to E4) facilitate extensive modulation of the host cell transcriptional machinery (E1, which is often considered as two regions [E1A and E1B] and E4), comprise the virus DNA replication complex (E2), and provide means for subverting host defence mechanisms (E3). E2 is well-conserved throughout the family, whereas the length and gene content of E1, E3 and E4 exhibit great variability even within genera (Figure 2 Adenoviridae). Intermediate proteins (IX and IVa2) and late (L) gene products (L1–L5) are concerned with virion assembly and maturation (Ahi and Mittal 2016, Ahi et al., 2017).
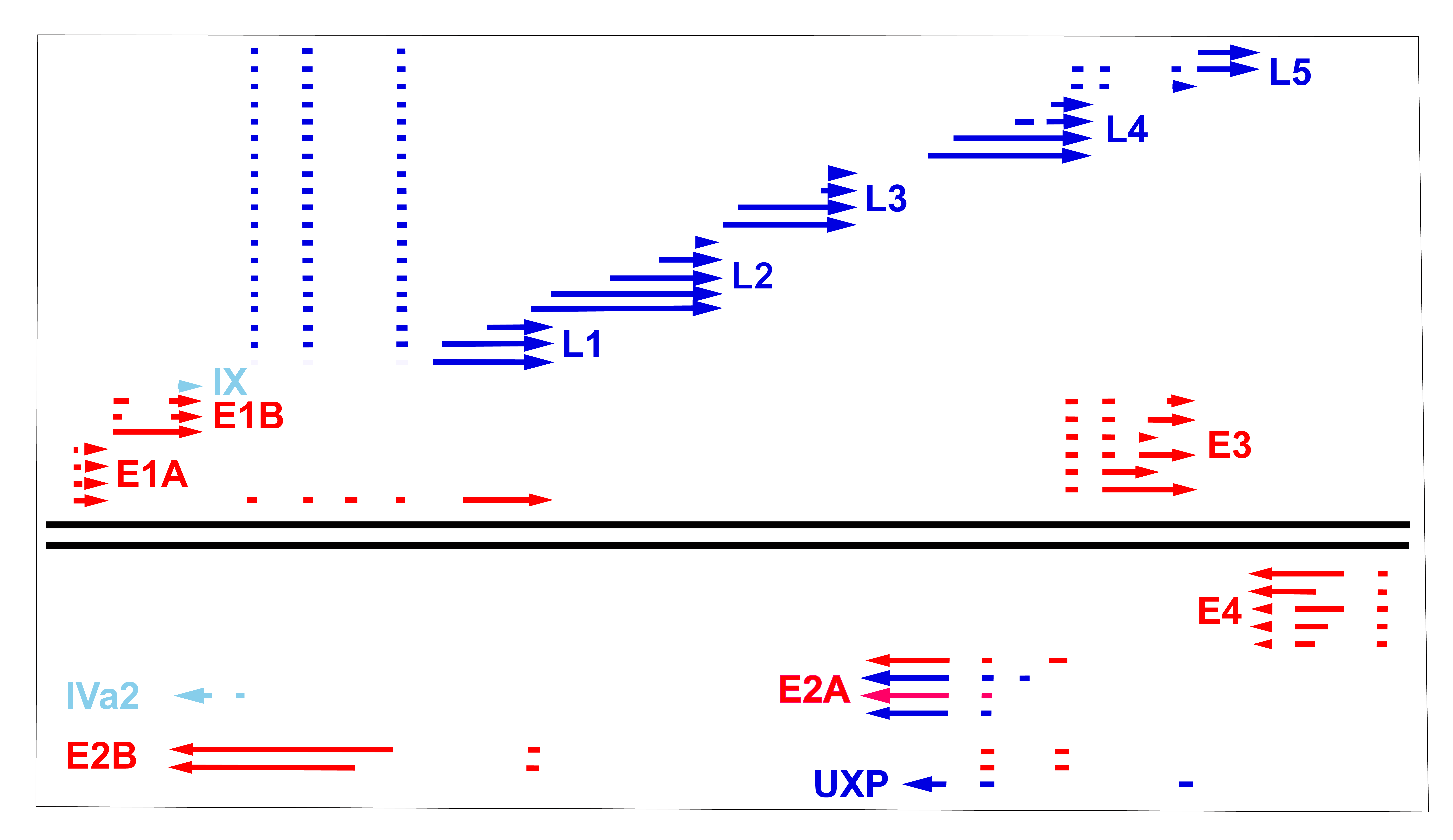 |
| Figure 3 Adenoviridae Schematic illustration of the transcription pattern of human adenovirus 2. The parallel lines indicate the linear dsDNA genome of 36 kb. The dots, broken lines and split arrows indicate the spliced structures of the mRNAs. E1A, E1B, E2A, E2B, E3 and E4 refer to early transcription units (red arrows). Most (but not all) late genes (blue arrows) are in the major late transcription unit, which initiates after the E1B and protein IX genes on the rightward transcribed strand, and which includes the L1, L2, L3, L4 and L5 families of mRNAs. Intermediate genes (encoding protein IX and protein IVa2) are shown by light blue arrows. (Adapted from Wold and Gooding 1991). |
Biology
The natural host range of adenoviruses has been thought to be confined to a single host species (e.g. members of the species Mastadenovirus dominans are restricted to humans), or to evolutionarily closely related species (e.g. bovine adenovirus 2, which is confined to cattle and sheep). This also applies to cells in culture. However, there is now evidence that numerous adenoviruses can cross host species barriers. An extreme example is skunk adenovirus 1 (Mastadenovirus trianonense), which was first described in Canada and has also been detected in diseased or dead African pigmy hedgehogs in Japan and the USA, North American porcupines, grey fox, racoon, domestic ferret and even in a New World monkey in a Hungarian zoo (Kozak et al., 2015, Madarame et al., 2019, Needle et al., 2019a, Balik et al., 2020, Doszpoly et al., 2020, Needle et al., 2020, Bourque et al., 2022, Orbay-Cerrato et al., 2024) and these are candidates for classification. Furthermore, according to its phylogenetic relationships and the arrangement of genes in the E3 region, this virus may have originated from a bat adenovirus. Similarly, canine adenovirus 1 (CAdV-1) has been detected not only in dogs but also in multiple carnivore species, wolves, various fox species, raccoons, bears and skunks (Balboni et al., 2019). A broader host range is often observed in members of genera Barthadenovirus and Siadenovirus, where a host switch may have played an important role in the evolutionary history. For example, the barthadenovirus snake adenovirus 1 occurs in corn snake, boa, python and eastern indigo snake (Benkő et al., 2002, Farkas et al., 2008, Bogan et al., 2024), and snake adenovirus 2 has been detected in eastern corn snake, California kingsnake and asp viper (Garner et al., 2008). Furthermore, both of these snake adenovirus types occur in tortoises (Salzmann et al., 2021). The siadenovirus psittacine adenovirus 2 has been described in representatives of a dozen species of the order Psittaciformes (Wellehan et al., 2009, Ballmann and Vidovszky 2013, Phalen et al., 2019, Yang et al., 2019, Zadravec et al., 2022), whereas the siadenovirus raptor adenovirus 1 has been found in Harris’s hawk and two owl species (Zsivanovits et al., 2006, Kovács and Benkő 2011). Some adenoviruses infecting birds are reported in several avian species with infections ranging from non-symptomatic to those with severe clinical signs (Athukorala et al., 2022, Schachner and Hess 2024).
Some HAdVs, mainly members of the species Mastadenovirus caesari, productively infect rodent or ruminant cells. HAdVs of species Mastadenovirus adami and Mastadenovirus blackbeardi manifest high and low oncogenicity, respectively, in newborn hamsters, whereas the other HAdVs are not oncogenic. The majority of HAdV infections in humans are subclinical but can result in virus shedding for years due to persistent infection of lymphocytes. Direct or indirect transmission occurs from throat, faeces, eye, urine or fomites, depending on the virus type. Certain HAdV types (listed below in parentheses) are predominantly associated with specific pathology, such as acute respiratory infections (HAdV-1 to -5, -7, -14, -21) (Robinson et al., 2011, Hage et al., 2014, Kajon et al., 2019), adenoidal–pharyngeal conjunctivitis (HAdV-3, -4, -7, -14), epidemic keratoconjunctivitis (HAdV-8, -19, -37, -53, -54, -56) (Walsh et al., 2009, Aoki et al., 2019), hepatitis (HAdV-5) or venereal disease (HAdV-37, -56, -64) (Hanaoka et al., 2019). HAdV-40 and HAdV-41 can be detected in high yield from faeces of young children with acute gastroenteritis and are second only to rotaviruses as a major cause of infantile viral diarrhoea (Bates et al., 1993). HAdV-11, -34 and -35 cause persistent interstitial infection in the kidney and haemorrhagic cystitis, occurring most frequently in immuno-suppressed patients after stem cell or solid organ transplantation. The most severe and frequently lethal infections in primary immunodeficiencies and paediatric allogeneic hematopoietic stem cell transplantation are caused by members of species Mastadenovirus adami, Mastadenovirus blackbeardi and Mastadenovirus caesari; HAdV-31 (Mastadenovirus caesari) being most common. Patients with secondary immunodeficiency can also be at risk when infected by members of these three species, which can cause different manifestations of disseminated disease. Fatal liver disease can develop after infection with HAdV-1, -2 or -5 (Lion 2014). Serotypes from HAdV-42 to HAdV-49, and HAdV-51 of species Mastadenovirus dominans have all been isolated from AIDS patients but do not cause severe disease (De Jong et al., 1999, Al Qurashi et al., 2012).
Adenoviruses infecting susceptible cells cause similar cytopathic effects, i.e., early rounding of cells and aggregation or lysis of chromatin, followed by the later appearance of characteristic eosinophilic or basophilic nuclear inclusions.
Mastadenovirus infections are common in other mammals, but disease usually appears only if predisposing factors (e.g. management problems, crowding, shipping or concurrent bacterial infections) are present. CAdV-1 seems to be an exception, in that it is the causative agent of infectious canine hepatitis (Rubarth's disease), a life-threatening disease of puppies, and of encephalitis in foxes. A closely related virus, canine adenovirus 2 (CAdV-2), causes infectious laryngotracheitis (kennel cough) in dogs, and this disease is common in breeder stocks. Squirrel monkey adenoviruses were described from fatal infections during immunosuppression (Rogers et al., 2020).
Hepatitis-hydropericardium syndrome predominantly in chickens has been associated with fowl adenovirus 4 (FAdV-4). This disease occurs mainly in Asia and causes great economic losses. Gizzard erosion in chickens is caused by fowl adenovirus 1, whereas inclusion body hepatitis is caused by fowl adenovirus 2, fowl adenovirus 8a, fowl adenovirus 8b or fowl adenovirus 11 in various bird species (Schachner et al., 2018). Duck adenovirus 1 is the causative agent of egg drop syndrome of chickens (Hess et al., 1997). The disease causes dramatic decrease in the egg production of laying flocks. Turkey adenovirus 3 (genus Siadenovirus) causes turkey haemorrhagic enteritis (Suresh and Sharma 1996, Pitcovski et al., 1998, Palya et al., 2007).
HAdV-5 has been engineered and used extensively as a gene delivery vector (Alonso-Padilla et al., 2016). Other serotypes (including non-human ones) are being developed to overcome the problem posed by pre-existing neutralizing antibodies in the population, and also to achieve better targeting of specific organs and tissues (Both 2004, Dicks et al., 2012, Abbink et al., 2015, Junyent and Kremer 2015, Alonso-Padilla et al., 2016, Corredor et al., 2017, Del Rio et al., 2019, Sayedahmed et al., 2019, Lu et al., 2020, Alhashimi et al., 2021). Recently, chimpanzee adenovirus-based vector vaccines have been developed against very important human pathogens, such as Ebola virus (Filoviridae) and SARS-CoV-2 (Coronaviridae) (Ledgerwood et al., 2017, Folegatti et al., 2020). Adenoviruses have been tested as the basis of direct oncolytic treatment (Bots et al., 2024).
Antigenicity
In the past, adenovirus serotypes were differentiated based on cross-neutralization assays (Mennechet et al., 2019). A serotype is defined as an adenovirus that either exhibits no cross-reaction with other adenoviruses or shows a homologous:heterologous titer ratio greater than 16 (in both directions). For homologous:heterologous titer ratios of 8 or 16, a serotype assignment is made if either the viral hemagglutinins are unrelated (as shown by lack of cross-reaction in hemagglutination-inhibition tests) or if substantial biophysical, biochemical or phylogenetic differences exist. Antigens at the surface of the virion are mainly type-specific. Hexons are involved in neutralization, and fibers in neutralization and hemagglutination-inhibition. Soluble antigens associated with virus infections include surplus capsid proteins that have not been assembled. As defined using monoclonal antibodies, hexons and other soluble antigens carry numerous epitopes that can be genus-, species- or type-specific. Free hexon protein reacts mainly as a genus-specific antigen. The genus-specific antigen is located on the basal surface of the hexon, whereas serotype-specific antigens are located mainly on the tower region (Adám et al., 1998).
Genus demarcation criteria
Genus designation depends on the following characteristics:
- Phylogenetic distance (based primarily on maximum likelihood analysis of the pol amino acid sequence)
- Genome organization (presence or lack of certain characteristic genes or transcription units)
- Host range
Derivation of names
The adenovirus species have Latinized binomial names. The Latinised nomenclature was developed in accordance with the guidelines established by (Postler et al., 2022) 36748479.
Adenoviridae: from the Ancient Greek ἡ ἀδήν (aden), meaning “gland”; in recognition of the fact that adenoviruses were first isolated from human adenoid tissue.
Aviadenovirus: from the Latin avis, meaning “bird”.
Aviadenovirus anatidae: from the family name Anatidae (duck AdV-5 isolated from wild duck, Anas platyrhynchos, and Pacific black duck, A. superciliosa).
Aviadenovirus anatis (earlier Duck aviadenovirus B): from the Latin anas, meaning “duck” (Muscovy duck).
Aviadenovirus anseris (earlier Goose aviadenovirus A): from the Latin anser, meaning “goose”.
Aviadenovirus bubonis: from the Latin bubo, meaning “owl” (Indian eagle-owl, Bubo bengalensis).
Aviadenovirus cairinae: from genus name of Cairina moschata (duck AdV-4, Muscovy duck).
Aviadenovirus columbae (earlier Pigeon aviadenovirus A): from the Latin columba, meaning “pigeon” (pigeon AdV-1).
Aviadenovirus columbidae (earlier Pigeon aviadenovirus B): from the family name Columbidae (pigeon AdV-2).
Aviadenovirus falconis (earlier Falcon aviadenovirus A): from the Latin falco, meaning “falcon” (several falcon species).
Aviadenovirus gallinae (earlier Fowl aviadenovirus D): from the Latin gallina, meaning “hen” (fowl AdV-2, -3, -9, -11).
Aviadenovirus gallopavoprimum (earlier Turkey aviadenovirus B): from the species epithet gallopavo and the Latin primum, meaning “first” (referring to turkey AdV-1).
Aviadenovirus gallopavoquartum (earlier Turkey aviadenovirus C): from the species epithet gallopavo and the Latin quartum, meaning “fourth” (referring to turkey AdV-4).
Aviadenovirus gallopavoquintum (Turkey aviadenovirus D): from the species epithet gallopavo and the Latin quintum, meaning “fifth” (referring to turkey AdV-5).
Aviadenovirus gruis: from the Latin grus, meaning “crane”.
Aviadenovirus hepatitidis (earlier Fowl aviadenovirus E): from the Latin disease name hepatitis referring to inclusion body hepatitis in chickens (fowl AdV-6, -7, -8a, -8b).
Aviadenovirus hydropericardii (earlier Fowl aviadenovirus C): from the Latin disease name hydropericardium syndrome caused by fowl AdV-4 in chickens (additional member: fowl AdV-10).
Aviadenovirus leukophtalmi: from the Latin species epithet of Psittacara leucophthalmus, referring to white-eyed parakeet (white-eyed parakeet adenovirus 2).
Aviadenovirus podargidae: from the family name Podargidae, which includes the frogmouth (bird).
Aviadenovirus quintum (earlier Fowl aviadenovirus B): from the Latin quintum, meaning “fifth” (referring to fowl AdV-5).
Aviadenovirus rubri (earlier Psittacine aviadenovirus B): from the Latin rubrum, meaning “red” (red-bellied parrot, Poicephalus rufiventris, psittacine AdV-4).
Aviadenovirus senegalense: (earlier Psittacine aviadenovirus C): from the species epithet of Poicephalus senegalus (Senegal parrot, psittacine AdV-1).
Aviadenovirus spinus: from the species epithet of Spinus spinus (Eurasian siskin).
Aviadenovirus turdi: from the Latin turdus, meaning “thrush”.
Aviadenovirus ventriculi (earlier Fowl aviadenovirus A): from the Latin ventriculus, meaning “gizzard” and referring to the gizzard erosion in chickens and quails caused by fowl AdV-1.
Barthadenovirus (earlier Atadenovirus): from the name of Hungarian scientist Adorján Bartha, who was the first to recognize the existence of a specific adenovirus lineage in cattle.
Barthadenovirus amazonae (earlier Psittacine atadenovirus A): from the genus name Amazona farisona (Southern mealy amazon).
Barthadenovirus bosquartum (earlier Bovine atadenovirus D): from the Latin bos, meaning “cow”, and the Latin quartus, meaning “fourth” referring to bovine AdV-4.
Barthadenovirus bosseptimum: from the Latin bos, meaning “cow”, and the Latin septimum, meaning “seventh” referring to bovine AdV-7.
Barthadenovirus bossextum (earlier Bovine atadenovirus E): from the Latin bos, meaning “cow”, and the Latin sextum, meaning “sixth” referring to bovine AdV-6.
Barthadenovirus caerulei: from the Latin caeruleum, meaning “blue” (blue-throated macaw, psittacine AdV-11).
Barthadenovirus cervi (earlier Deer atadenovirus A): from the Latin cervus, meaning “deer” (deer AdV-1, syn. Odocoileus AdV-1).
Barthadenovirus draconis (earlier Lizard atadenovirus B): from the Latin draco, meaning “dragon” (bearded dragon).
Barthadenovirus galloanserae (earlier Duck atadenovirus A): from the superorder name Galloanserae (chicken, waterfowl; duck AdV-1).
Barthadenovirus inornati: from the species epithet of Phylloscopus inornatus, meaning “plain” (yellow-browed warbler, Passeriformes).
Barthadenovirus lacertae (earlier Lizard atadenovirus B): from the Latin lacerta, meaning “lizard” (Mexican beaded lizard).
Barthadenovirus macropodidae: from the family name Macropodidae, a family of marsupials (agile wallaby).
Barthadenovirus mellis: from the Latin mel, meaning “honey” (eastern spinebill, a honeyeater; Passeriformes).
Barthadenovirus ovis (earlier Ovine atadenovirus D): from the Latin ovis, meaning “sheep” (ovine AdV-7).
Barthadenovirus schwarzi: from the species epithet of Phylloscopus schwarzi, referring to German astronomer Ludwig Schwarz (Radde's warbler, Passeriformes).
Barthadenovirus serpentis (earlier: Snake atadenovirus A): from the Latin serpens, meaning “snake” (snake AdV-1).
Barthadenovirus sternae: from the genus name Sterna hirundo (common tern).
Barthadenovirus vulpeculae (earlier: Possum atadenovirus A): from the Latin species epithet of Trichosurus vulpecula, meaning “little fox" (common brushtail possum).
Ichtadenovirus: from the Ancient Greek ὁ ἰχθῦς (ichthys), meaning “fish”.
Ichtadenovirus acipenseris (earlier Sturgeon ichtadenovirus A): from the Latin acipenser, meaning “sturgeon” (white sturgeon, Acipenser transmontanus).
Mastadenovirus: from the Greek ὁ μαστός (mastos), meaning “breast” referring to the mammalian hosts of viruses in this genus.
Mastadenovirus adami (earlier Human mastadenovirus A): from Adam, the first man (referring to the letter A in the earlier species name).
Mastadenovirus aegyptiaci (earlier Bat mastadenovirus I): from the species epithet Rousettus aegyptiacus (Egyptian fruit bat).
Mastadenovirus alienum (earlier: Simian mastadenovirus E): from the Latin alienus, meaning “foreign, strange” (because simian AdV-16 has unique hemagglutination properties: type IV).
Mastadenovirus asiense (earlier Bat mastadenovirus J): from the Latin Asia, meaning “Asia”, referring to the name of the continent in the English name of the host: Asian parti-colored bat (Vespertilio sinensis, isolate Vs9)
Mastadenovirus blackbeardi (earlier Human mastadenovirus B): from the name Blackbeard (the pirate, because viruses act as pirates; referring to the letter B in the earlier species name).
Mastadenovirus bosdecimum (earlier: Bovine mastadenovirus C): from the Latin bos, meaning “cow”, and the Latin decimum, meaning “tenth” (referring to bovine AdV-10).
Mastadenovirus bosprimum (earlier Bovine mastadenovirus A): from the Latin bos, meaning “cow”, and the Latin primum, meaning “first” (referring to bovine AdV-1).
Mastadenovirus bostertium (earlier: Bovine mastadenovirus B): from the Latin bos, meaning “cow”, and the Latin tertium, meaning “third” (referring to bovine AdV-3).
Mastadenovirus bovidae (earlier Ovine mastadenovirus A): from the family name Bovidae, referring to the fact that its members are found both in cattle and sheep (bovine AdV-2, ovine AdV-2 to 4).
Mastadenovirus caesari (earlier Human mastadenovirus C): from the Latin name Caesar referring to the letter C in the earlier species name (because HAdV-2 and HAdV-5 “rule” like an emperor in almost all adenovirology laboratories).
Mastadenovirus canidae (earlier Canine mastadenovirus A): from the family name Canidae (dog, wolf, various foxes, etc.).
Mastadenovirus caviae (earlier Guinea pig mastadenovirus A): from the genus name of Cavia porcellus (guinea pig).
Mastadenovirus cervi (earlier Deer mastadenovirus B): from the Latin cervus, meaning “deer” (deer AdV-2).
Mastadenovirus chalinolobi: from the genus name of Chalinolobus gouldii (Gould's wattled bat; lobus meaning “lobe”, referring to the fleshy lobes at the bottom edge of the ears and on the lower lips).
Mastadenovirus chlorocebi (earlier: Simian mastadenovirus F): from the genus name of Chlorocebus aethiops (grivet, simian AdV-17, -18).
Mastadenovirus cordis (earlier Murine mastadenovirus C): from the Latin cor, meaning “heart”, referring to the cardiotropic property of murine AdV-3 (from striped field mouse, Apodemus agrarius).
Mastadenovirus cynocephali (earlier: Simian mastadenovirus C): from the species epithet of Papio cynocephalus, meaning "dog-faced" (yellow baboon, simian AdV-19).
Mastadenovirus delphini (earlier Dolphin mastadenovirus B): from the Latin delphinus, meaning “dolphin” (common bottlenose dolphin, dolphin AdV-1).
Mastadenovirus delphinidae (earlier Dolphin mastadenovirus A): from the family name of Delphinidae, which includes common bottlenose dolphin (dolphin AdV-2).
Mastadenovirus dipodomysis: from the genus name of Dipodomys ordii (Ord’s kangaroo rat, Rodentia).
Mastadenovirus dominans (earlier Human mastadenovirus D): from the Latin dominans, meaning “dominating, most abundant” (referring to the highest number of known human AdV serotypes and to the letter D in the earlier species name).
Mastadenovirus eidoli (earlier Bat mastadenovirus H): from the genus name of Eidolon helvum (straw-colored fruit bat).
Mastadenovirus encephalomyelitidis (earlier Murine mastadenovirus A): from the Latin name of the disease encephalomyelitis, referring to the fact that murine AdV-1 can cause fatal encephalomyelitis.
Mastadenovirus equi (earlier Equine mastadenovirus A): from the Latin equus, meaning “horse” (equine AdV-1; host switch from bat; pathogenic).
Mastadenovirus equidae (earlier Equine mastadenovirus B): from the family name Equidae, which includes horses (equine AdV-2, coevolved with horses).
Mastadenovirus exoticum (earlier Human mastadenovirus E) from the Latin exoticum, meaning “exotic”, referring that it contains an exotic single human type among chimpanzee AdV types (HAdV-4; referring to the letter E in the earlier species name).
Mastadenovirus flavi (earlier Smian mastadenovirus I): from the Latin flavus, meaning “yellow” referring to “golden” snub-nosed monkey (golden-haired).
Mastadenovirus faecale (earlier Human mastadenovirus F) from the Latin faex, meaning “dregs, sediment”, referring that HAdV-40 and HAdV-41 are detectable mainly in faeces (referring to the letter in the old species name).
Mastadenovirus humile (earlier Bat mastadenovirus E): from the Latin humilis, meaning ”low” referring to the lowest known G+C composition among AdVs (bat AdV-8 isolate WIV13).
Mastadenovirus lamiae: from the Latin lamia, meaning “witch” (grey mouse lemur, reddish-grey mouse lemur; lemurs: from the Latin lemures, meaning “ghosts” or “spirits”).
Mastadenovirus longumcaudae (earlier Simian mastadenovirus B): from Latin longus, meaning “long” and cauda, meaning “(animal) tail” (several AdV types from crab-eating or long-tailed macaque, Macaca fascicularis).
Mastadenovirus macacae (earlier Simian mastadenovirus D): from the genus of Macaca sp. (macaque, SAdV-13).
Mastadenovirus magnauris (earlier Bat mastadenovirus G): from the Latin magnus, meaning “big” and auris, meaning “ear” (Rafinesque's big-eared bat, Corynorhinus rafinesquii, bat AdV-11).
Mastadenovirus ovisoctavum (earlier Ovine mastadenovirus C): from the Latin ovis, meaning “sheep”, and the Latin octavum, meaning “eighth” (referring to ovine AdV-8).
Mastadenovirus ovisprimum (earlier Ovine mastadenovirus B): from the Latin ovis, meaning sheep, and the Latin primum, meaning “first” (referring to ovine AdV-1).
Mastadenovirus miniopteridae (earlier Bat mastadenovirus D): from the family name of Miniopterus schreibersi (bat AdV-7 isolate WIV12).
Mastadenovirus muris (earlier Murine mastadenovirus B): from the Latin mus, meaning “mouse” (murine AdV-2, numerous virus variants reflecting a long coevolution).
Mastadenovirus musauriti (earlier Bat mastadenovirus A): from the Latin mus, meaning “mouse”, and the Latin auritus, meaning “with or having ears” referring to mouse-eared bats (the genus Myotis, a neo-Latin construction of ”mouse-eared” from the Greek ὁ μῦς (mys), meaning "mouse” and τό οὖς (oûs), meaning “ear”; Myotis ricketti, bat AdV-3).
Mastadenovirus otariidae (earlier Sea lion mastadenovirus A): from the family name of Zalophus californianus (California sea lion).
Mastadenovirus phocoenae: from the genus name of Phocoena phocoena (harbor porpoise).
Mastadenovirus pipistrelli (earlier Bat mastadenovirus B): from the species epithet of Pipistrellus pipistrellus (common pipistrelle, bat AdV-2).
Mastadenovirus porcustertium (earlier Porcine mastadenovirus A): from the Latin porcus, meaning “swine”, and the Latin tertium, meaning “third” referring to porcine AdV-3 (but also including porcine AdV-1 and -2).
Mastadenovirus porcusquartum (earlier Porcine mastadenovirus B): from the Latin porcus, meaning swine, and the Latin quartum, meaning “fourth” (referring to porcine AdV-4).
Mastadenovirus porcusquintum (earlier Porcine mastadenovirus C): from the Latin porcus, meaning “swine”, and the Latin quintum, meaning “fifth” (referring to porcine AdV-5).
Mastadenovirus pteropodidae (earlier Bat mastadenovirus F): from the family name of Rousettus leschenaultii (bat AdV-9 isolate WIV17).
Mastadenovirus rhesi (earlier Simian mastadenovirus H): rhesus is reminiscent of the mythological king Rhesus of Thrace in the Iliad (rhesus macaque, Macaca mulatta, SAdV-54).
Mastadenovirus rhinolophidae (earlier Bat mastadenovirus C): from the family name of Miniopterus schreibersi (bat AdV-4) and Rhinolophus cornutus (bat AdV isolate Rc-kw20).
Mastadenovirus russelli (earlier Human mastadenovirus G): honouring the respected Scottish adenovirologist William C. Russell (HAdV-52 and numerous Old World monkey AdVs).
Mastadenovirus sciuri (earlier Squirrel mastadenovirus A): from the Latin sciurus, meaning “squirrel”.
Mastadenovirus simiae (earlier Simian mastadenovirus A): from the Latin simia, meaning “monkey” (Old World monkeys, e.g. simian AdV-3 from rhesus macaque).
Mastadenovirus simiavigesimum (earlier Simian mastadenovirus G): from the Latin simia, meaning “monkey”, and vigesimum, meaning “twentieth” (referring to simian AdV-20).
Mastadenovirus simuli (earlier Platyrrhini mastadenovirus A): from the Latin simulus, meaning "flat nosed", referring to New World monkeys (Platyrrhini) as Platyrrhini is derived from the Greek for "broad nosed", as the noses of these monkey are flatter than those of other simians (coper titi monkey, Plecturocebus cupreus; black-capped squirrel monkey, Saimiri boliviensis).
Mastadenovirus tarandri: from the Latin tarandrus, meaning “reindeer”.
Mastadenovirus trianonense (earlier Skunk mastadenovirus A): from name of the treaty of Trianon, referring to the fact that this virus is fatal and crosses (species) boundaries like the treaty (skunk AdV-1 described from skunk (Canada), found in four-toed hedgehog (Africa, Japan, USA), pygmy marmoset (South America, Hungary), North American porcupine, grey fox and raccoon (North America) – with a supposed bat AdV origin).
Mastadenovirus tupaiae (earlier Tree shrew mastadenovirus A): from the genus name of Tupaia (referring to Northern treeshrew, Tupaia belangeri).
Mastadenovirus ursi (earlier Polar bear mastadenovirus A): from the Latin ursus meaning “bear” (polar bear AdV-1).
Siadenovirus: from sialidase, in recognition that most members of the genus encode a putative sialidase homologue.
Siadenovirus carbocapituli: from the Latin carbo, meaning “coal” and the Latin caput, meaning “head” (referring to the Hungarian name széncinege of Parus major, which refers to a “coal-headed tit”; great tit AdV-3).
Siadenovirus cinerei: from the Latin cinereum, meaning “ash-coloured” (grey parrot, PsAdV-2)
Siadenovirus gallopavotertium (earlier Turkey siadenovirus A): from the species epithet gallopavo and the Latin tertium, meaning “third” (referring to turkey AdV-3).
Siadenovirus paridae (earlier Great tit siadenovirus A): after the family name of Paridae (great tit, blue tit; great tit AdV-1).
Siadenovirus ranae (earlier Frog siadenovirus A): from the Latin rana, meaning “frog” (frog AdV-1).
Siadenovirus raptoris (earlier Raptor siadenovirus A): from the Latin raptor, meaning “robber, plunderer” (Accipitriformes and Strigiformes; raptor AdV-1).
Siadenovirus sanguineae (earlier Psittacine siadenovirus E): after the species epithet of Cacatua sanguinea, meaning “red chalk/redddish-brown color/color of dried blood” (little corella, psittacine AdV-7).
Siadenovirus spheniscidae (earlier Penguin siadenovirus A): after the family name (chinstrap and some further penguin species).
Siadenovirus stercorariidae (earlier Skua siadenovirus A): after the family name of Stercorarius maccormicki (South Polar skua).
Siadenovirus uriae: after the genus name of Uria aalge (from ancient Greek ἡ οὐρία (ouria), a waterbird mentioned by Athenaeus; common murre, Charadriiformes).
Siadenovirus viridis (earlier Psittacine siadenovirus D): from the Latin viridis, meaning “green” (Pacific parrotlet, a small green parrotlet, psittacine AdV-5).
Testadenovirus: from the Latin testudo, meaning “tortoise”.
Testadenovirus trachemysis (earlier Pond slider testadenovirus A): from the genus name of Trachemys scripta (pond slider, red-eared slider AdV-1).
Relationships within the family
High-resolution genomic data have provided insights into the molecular evolution of adenoviruses. Consistent with specific characteristics of genome organization, molecular phylogenetic analysis results in clear separation of six different clusters corresponding to the six accepted genera (Mastadenovirus, Aviadenovirus, Barthadenovirus, Siadenovirus, Ichtadenovirus and Testadenovirus; Figure 4 Adenoviridae) (Doszpoly et al., 2019, Harrach et al., 2019). Within genera, phylogenetic relationships among the viruses are usually similar to those among their hosts, i.e., the tree topologies of the viruses and the hosts are often congruent (Benkő and Harrach 2003, Chen et al., 2011, Ballmann and Harrach 2016, Podgorski et al., 2018, Harrach et al., 2019, Kaján et al., 2020). An interesting example is a common clade of mastadenoviruses from animals belonging to the order Artiodactyla clade Cetruminantia (cetaceans and ruminants) (Standorf et al., 2018, Harrach et al., 2019, Vidovszky et al., 2022). Phylogenetic analyses have revealed separable phylogroups among isolates or strains of the same serotype, such as human adenovirus 4 (HAdV-4) (Gonzalez et al., 2019).
 |
| Figure 4 Adenoviridae Phylogenetic tree of representative members of adenovirus species based on maximum likelihood analysis of the complete amino acid sequences of DNA-dependent DNA polymerase. Complete pol amino acid sequences were aligned using the MAFFT v7.490 G-INS-i algorithm. The alignment was edited using the Automated1 algorithm of TrimAl (v1.3): the length of the edited alignment was 805 amino acids. The LG+I+G evolutionary model was selected using ModelTest-NG v0.1.7. The tree was inferred using RAxML-NG v1.2.2, and the robustness of the tree was determined with a non-parametric bootstrap calculation using 1000 replicates. The transfer bootstrap expectation values were applied to the tree. The phylogenetic tree was visualized using MEGA7, and it was rooted on the midpoint, with the bootstrap values ≥75% at the nodes. The bar indicates 20% difference between two neighbouring sequences. Viruses are represented by GenBank accession number, virus name with type or strain designation, and species name. Coloured spots designate exemplars of accepted species, coloured (empty) circles designate members of proposed species. This phylogenetic tree and corresponding sequence alignment are available to download from the Resources page. |
Against this coevolutionary background, there are some exceptional cases of very distantly related adenoviruses infecting the same host. In particular, the adenovirus types isolated from cattle, sheep, deer or birds appear on very distant branches, and even in separate clusters corresponding to different genera (Ballmann and Harrach 2016, Duarte et al., 2019, Harrach et al., 2019, Sutherland et al., 2019, Vidovszky et al., 2019). Thus, it appears that multiple host switches may also have occurred (Harrach et al., 2019). For example, it is probable that adenoviruses from scaled reptiles switched to ruminants and birds (Benkő and Harrach 2003, Wellehan et al., 2004), adenoviruses from bats switched to carnivores (causing fatal disease in dogs) (Kohl et al., 2012) and horses (Vidovszky et al., 2015), and adenoviruses from Old World monkeys and apes switched to humans (Hoppe et al., 2015). Duck adenovirus 1 (DAdV-1) switched from ducks to chickens.
Relationships with other taxa
The dsDNA bacteriophage Enterobacteria phage PRD1 (family Tectiviridae), shares a similar virion architecture (a pseudo T=25 icosahedral capsid with fiber-like projections at the vertices) with adenoviruses (Butcher et al., 1995). The 15 kb genome of PRD1 has ITRs and contains genes encoding TP and pol arranged in the same order as in adenoviruses. Moreover, the TP also acts as primer in PRD1 DNA replication. A study of the structure of the main capsid proteins (P3 of PRD1 and the hexon of adenoviruses) revealed a very similar fold (a double jelly roll orthogonal to the capsid surface) and further strengthened an evolutionary link between the two viruses (Benson et al., 1999). Such a double-jelly-roll fold has been found in many virus capsids (San Martín and van Raaij 2018).
In fungi and plants, a linear plasmid (called yeast killer plasmid) occurs either in the cytoplasm or in the mitochondria that has adenovirus-like features (an ITR and a TP gene adjacent to a pol gene). Based on the established or predicted three-dimensional structures of their major capsid proteins and on sharing of a characteristic ATPase, Sulfolobus turreted icosahedral virus (family Turriviridae), which infects a crenarchaeal host (in domain Archaea), and also two archaeal proviruses (TKV4 and MVV) could also be placed into the PRD1-adenovirus lineage (Krupovic and Bamford 2008a). These archaeal proviruses (TKV4 and MVV) are integrated into the 5′- and 3′-distal regions of tRNA genes of the euryarchaeal species Thermococcus kodakaraensis (strain KOD1) and Methanococcus voltae (A3), respectively. Recent studies have proposed the existence of novel virus families for adomaviruses and adintoviruses, which are related to adenoviruses on the basis of several homologous genes (Starrett et al., 2021).
Examination of the deepest phylogeny of the family suggests an origin from bacterial viruses by descent from non-replicating polintons in the mitochondria that became replicating adenoviruses in vertebrates only (Krupovic and Bamford 2008b, Koonin et al., 2015, Krupovic and Koonin 2015). These distant similarities, which are focused primarily on the structural conservation of part of the hexon in other viruses, led to the establishment of the following higher taxa incorporating the family Adenoviridae: realm Varidnaviria, kingdom Bamfordvirae (viruses with a double jelly-roll major capsid protein), phylum Preplasmaviricota, class Tectiliviricetes (to which also the family Tectiviridae belongs), order Rowavirales (where the family Adenoviridae is alone) (Koonin et al., 2020, Walker et al., 2020). A proposal has been submitted to ICTV suggesting the establishment of two subphyla in the phylum Preplasmaviricota, one named Prepoliviricotina for the Tectiliviricetes (with the single family of Tectiviridae, which are procaryote viruses), and Polisuviricotina for ten families of eukaryotic viruses including the Adenoviridae, which (alone) would get the newly proposed class name Pharingeaviricetes (https://ictv.global/filebrowser/download/17320, https://ictv.global/filebrowser/download/17314) (Koonin et al., 2024).
Adenoviruses also show some similarity to other viruses. The fibers of many adenovirus types use the same cellular receptor (coxsackievirus and adenovirus receptor, CAR) for attachment as coxsackie B viruses (family Picornaviridae) (Arnberg 2012). Adenovirus fibers have been reported to show structural similarity to reovirus attachment protein sigma 1, which binds the junction adhesion molecule (JAM) receptor (Chappell et al., 2002). Adenoviruses may occur together with adenovirus-dependent parvoviruses, for which they may provide helper functions (Pénzes et al., 2020a). Similarity was observed between certain proteins coded by the E3 region of HAdVs and the RL11 gene family of human cytomegalovirus (family Orthoherpesviridae) (Davison et al., 2003a). The primary structure of the p32K protein, which is characteristic of barthadenoviruses, has similarity to bacterial small acid-soluble proteins (SASPs) commonly found in various spore-forming bacteria (Élő et al., 2003). The barthadenovirus LH3 capsid protein has a beta-helix fold typical of receptor binding spikes in tailed bacteriophages (Menéndez-Conejero et al., 2017).
Related, unclassified viruses
| Virus name | Accession number | Abbreviation |
| crocodile adenovirus 1 | ||
| dark-spotted frog (Pelophylax) adenovirus 1 (from brain) | PP352022 | |
| dark-spotted frog (Pelophylax) adenovirus 2 (from intestine) | PP352021; PP352023 | |
| New Zealand smelt (cucumber fish, Retropinna) adenovirus 1 | OR270034; OR270035; OR270037; OR270090 | |
| New Zealand smelt (cucumber fish, Retropinna) adenovirus 2 | OR270036; OR270092 | |
| platypus adenovirus 1 | BK066738 |
Virus names and virus abbreviations are not official ICTV designations.
Adenovirus-caused hepatitis has been described in crocodile hatchlings of about 2 weeks old that were bred on a commercial farm in South Africa (Pfitzer et al., 2019). The hatchlings showed typical clinical signs of hepatitis, and the identification of intranuclear inclusion bodies in the liver was used to differentiate between adenovirus-caused hepatitis and chlamydial hepatitis. However, no sequence data are available. Metagenomics revealed adenovirus sequences from cucumber fish (Retropinna retropinna), dark-spotted frog (Pelophylas nigromaculatus), and platypus (Ornithorhychus anatinus) (Grimwood et al., 2023, Buck et al., 2024). The recovered partial sequences differed from the known adenovirus sequences to an extent that these adenoviruses may merit the establishment of novel genera when full genomes will be known.

